Bioinformatic prediction and experimental verification of Fur-regulated genes in the extreme acidophile Acidithiobacillus ferrooxidans
- PMID: 17355989
- PMCID: PMC1874648
- DOI: 10.1093/nar/gkm068
Bioinformatic prediction and experimental verification of Fur-regulated genes in the extreme acidophile Acidithiobacillus ferrooxidans
Abstract
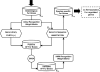
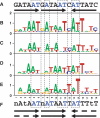
The gamma-proteobacterium Acidithiobacillus ferrooxidans lives in extremely acidic conditions (pH 2) and, unlike most organisms, is confronted with an abundant supply of soluble iron. It is also unusual in that it oxidizes iron as an energy source. Consequently, it faces the challenging dual problems of (i) maintaining intracellular iron homeostasis when confronted with extremely high environmental loads of iron and (ii) of regulating the use of iron both as an energy source and as a metabolic micronutrient. A combined bioinformatic and experimental approach was undertaken to identify Fur regulatory sites in the genome of A. ferrooxidans and to gain insight into the constitution of its Fur regulon. Fur regulatory targets associated with a variety of cellular functions including metal trafficking (e.g. feoPABC, tdr, tonBexbBD, copB, cdf), utilization (e.g. fdx, nif), transcriptional regulation (e.g. phoB, irr, iscR) and redox balance (grx, trx, gst) were identified. Selected predicted Fur regulatory sites were confirmed by FURTA, EMSA and in vitro transcription analyses. This study provides the first model for a Fur-binding site consensus sequence in an acidophilic iron-oxidizing microorganism and lays the foundation for future studies aimed at deepening our understanding of the regulatory networks that control iron uptake, homeostasis and oxidation in extreme acidophiles.
Figures



Similar articles
-
Microbial iron management mechanisms in extremely acidic environments: comparative genomics evidence for diversity and versatility.BMC Microbiol. 2008 Nov 24;8:203. doi: 10.1186/1471-2180-8-203. BMC Microbiol. 2008. PMID: 19025650 Free PMC article.
-
α-fur, an antisense RNA gene to fur in the extreme acidophile Acidithiobacillus ferrooxidans.Microbiology (Reading). 2014 Mar;160(Pt 3):514-524. doi: 10.1099/mic.0.073171-0. Epub 2014 Jan 2. Microbiology (Reading). 2014. PMID: 24385477
-
The ferric iron uptake regulator (Fur) from the extreme acidophile Acidithiobacillus ferrooxidans.Microbiology (Reading). 2005 Jun;151(Pt 6):2005-2015. doi: 10.1099/mic.0.27581-0. Microbiology (Reading). 2005. PMID: 15942007
-
Genomic insights into microbial iron oxidation and iron uptake strategies in extremely acidic environments.Environ Microbiol. 2012 Jul;14(7):1597-611. doi: 10.1111/j.1462-2920.2011.02626.x. Epub 2011 Nov 3. Environ Microbiol. 2012. PMID: 22050575 Review.
-
Genomic insights into the iron uptake mechanisms of the biomining microorganism Acidithiobacillus ferrooxidans.J Ind Microbiol Biotechnol. 2005 Dec;32(11-12):606-14. doi: 10.1007/s10295-005-0233-2. Epub 2005 May 14. J Ind Microbiol Biotechnol. 2005. PMID: 15895264 Review.
Cited by
-
Cobalamin Protection against Oxidative Stress in the Acidophilic Iron-oxidizing Bacterium Leptospirillum Group II CF-1.Front Microbiol. 2016 May 23;7:748. doi: 10.3389/fmicb.2016.00748. eCollection 2016. Front Microbiol. 2016. PMID: 27242761 Free PMC article.
-
Role of a Fur homolog in iron metabolism in Nitrosomonas europaea.BMC Microbiol. 2011 Feb 21;11:37. doi: 10.1186/1471-2180-11-37. BMC Microbiol. 2011. PMID: 21338516 Free PMC article.
-
Fox Cluster determinants for iron biooxidation in the extremely thermoacidophilic Sulfolobaceae.Environ Microbiol. 2022 Feb;24(2):850-865. doi: 10.1111/1462-2920.15727. Epub 2021 Aug 30. Environ Microbiol. 2022. PMID: 34406696 Free PMC article.
-
Bioinformatics in Latin America and SoIBio impact, a tale of spin-off and expansion around genomes and protein structures.Brief Bioinform. 2019 Mar 22;20(2):390-397. doi: 10.1093/bib/bbx064. Brief Bioinform. 2019. PMID: 28981567 Free PMC article.
-
Sorting out the mix in microbial genomics.Environ Microbiol. 2008 Dec;10(12):3187-92. doi: 10.1111/j.1462-2920.2008.01811.x. Environ Microbiol. 2008. PMID: 19025554 Free PMC article. No abstract available.
References
-
- Baichoo N, Wang T, Ye R, Helmann JD. Global analysis of the Bacillus subtilis Fur regulon and the iron starvation stimulon. Mol. Microbiol. 2002;45:1613–1629. - PubMed
-
- Hantke K. Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 2001;4:172–177. - PubMed
-
- McHugh JP, Rodríguez-Quiñones F, Abdul-Tehrani H, Svistunenko DA, Poole RK, Cooper CE, Andrews SC. Global iron-dependent gene regulation in Escherichia coli. A new mechanism for iron homeostasis. J. Biol. Chem. 2003;278:29478–29486. - PubMed
-
- Ochsner UA, Wilderman PJ, Vasil AI, Vasil ML. GeneChipR expression analysis of the iron starvation response in Pseudomonas aeruginosa: identification of novel pyoverdine biosynthesis genes. Mol. Microbiol. 2002;45:1277–1287. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

