In vivo and in vitro investigation of bacterial type B RNase P interaction with tRNA 3'-CCA
- PMID: 17355991
- PMCID: PMC1874595
- DOI: 10.1093/nar/gkm005
In vivo and in vitro investigation of bacterial type B RNase P interaction with tRNA 3'-CCA
Abstract
For catalysis by bacterial type B RNase P, the importance of a specific interaction with p(recursor)tRNA 3'-CCA termini is yet unclear. We show that mutation of one of the two G residues assumed to interact with 3'-CCA in type B RNase P RNAs inhibits cell growth, but cell viability is at least partially restored at increased RNase P levels due to RNase P protein overexpression. The in vivo defects of the mutant enzymes correlated with an enzyme defect at low Mg(2+) in vitro. For Bacillus subtilis RNase P, an isosteric C259-G(74) bp fully and a C258-G(75) bp slightly rescued catalytic proficiency, demonstrating Watson-Crick base pairing to tRNA 3'-CCA but also emphasizing the importance of the base identity of the 5'-proximal G residue (G258). We infer the defect of the mutant enzymes to primarily lie in the recruitment of catalytically relevant Mg(2+), with a possible contribution from altered RNA folding. Although with reduced efficiency, B. subtilis RNase P is able to cleave CCA-less ptRNAs in vitro and in vivo. We conclude that the observed in vivo defects upon disruption of the CCA interaction are either due to a global deceleration in ptRNA maturation or severe inhibition of 5'-maturation for a ptRNA subset.
Figures

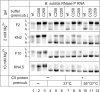
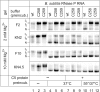
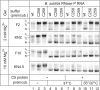

Similar articles
-
The precursor tRNA 3'-CCA interaction with Escherichia coli RNase P RNA is essential for catalysis by RNase P in vivo.RNA. 2006 Dec;12(12):2135-48. doi: 10.1261/rna.188306. Epub 2006 Oct 24. RNA. 2006. PMID: 17135488 Free PMC article.
-
Role of metal ions in the hydrolysis reaction catalyzed by RNase P RNA from Bacillus subtilis.J Mol Biol. 1999 Jul 9;290(2):433-45. doi: 10.1006/jmbi.1999.2890. J Mol Biol. 1999. PMID: 10390342
-
RNase P of the Cyanophora paradoxa cyanelle: a plastid ribozyme.Biochimie. 2007 Dec;89(12):1528-38. doi: 10.1016/j.biochi.2007.08.004. Epub 2007 Aug 11. Biochimie. 2007. PMID: 17881113
-
Structure of ribonuclease P--a universal ribozyme.Curr Opin Struct Biol. 2006 Jun;16(3):327-35. doi: 10.1016/j.sbi.2006.04.002. Epub 2006 May 2. Curr Opin Struct Biol. 2006. PMID: 16650980 Review.
-
Bacterial RNase P: a new view of an ancient enzyme.Nat Rev Microbiol. 2006 Oct;4(10):729-40. doi: 10.1038/nrmicro1491. Nat Rev Microbiol. 2006. PMID: 16980936 Review.
Cited by
-
RNase P Inhibitors Identified as Aggregators.Antimicrob Agents Chemother. 2021 Jul 16;65(8):e0030021. doi: 10.1128/AAC.00300-21. Epub 2021 Jul 16. Antimicrob Agents Chemother. 2021. PMID: 33972249 Free PMC article.
-
The contribution of the C5 protein subunit of Escherichia coli ribonuclease P to specificity for precursor tRNA is modulated by proximal 5' leader sequences.RNA. 2017 Oct;23(10):1502-1511. doi: 10.1261/rna.056408.116. Epub 2017 Jul 10. RNA. 2017. PMID: 28694328 Free PMC article.
-
Suppression of the Escherichia coli rnpA49 conditionally lethal phenotype by different compensatory mutations.RNA. 2024 Jul 16;30(8):977-991. doi: 10.1261/rna.079909.123. RNA. 2024. PMID: 38688559 Free PMC article.
-
Divergent molecular assembly and catalytic mechanisms between bacterial and archaeal RNase P in pre-tRNA cleavage.Proc Natl Acad Sci U S A. 2024 Oct 22;121(43):e2407579121. doi: 10.1073/pnas.2407579121. Epub 2024 Oct 16. Proc Natl Acad Sci U S A. 2024. PMID: 39413135 Free PMC article.
-
The Diversity of Ribonuclease P: Protein and RNA Catalysts with Analogous Biological Functions.Biomolecules. 2016 May 13;6(2):27. doi: 10.3390/biom6020027. Biomolecules. 2016. PMID: 27187488 Free PMC article. Review.
References
-
- Hartmann E, Hartmann RK. The enigma of ribonuclease P evolution. Trends Genet. 2003;19:561–569. - PubMed
-
- Evans D, Marquez SM, Pace NR. RNase P: interface of the RNA and protein worlds. Trends Biochem. Sci. 2006;31:333–341. - PubMed
-
- Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. - PubMed
-
- Schedl P, Primakoff P, Roberts J. Processing of E. coli tRNA precursors. Brookhaven Symp. Biol. 1974;26:53–76. - PubMed

