A novel microarray approach reveals new tissue-specific signatures of known and predicted mammalian microRNAs
- PMID: 17355992
- PMCID: PMC1874652
- DOI: 10.1093/nar/gkl1118
A novel microarray approach reveals new tissue-specific signatures of known and predicted mammalian microRNAs
Abstract
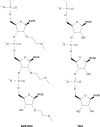
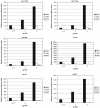
Microarrays to examine the global expression levels of microRNAs (miRNAs) in a systematic in-parallel manner have become important tools to help unravel the functions of miRNAs and to understand their roles in RNA-based regulation and their implications in human diseases. We have established a novel miRNA-specific microarray platform that enables the simultaneous expression analysis of both known and predicted miRNAs obtained from human or mouse origin. Chemically modified 2'-O-(2-methoxyethyl)-(MOE) oligoribonucleotide probes were arrayed onto Evanescent Resonance (ER) microchips by robotic spotting. Supplementing the complementary probes against miRNAs with carefully designed mismatch controls allowed for accurate sequence-specific determination of miRNA expression profiles obtained from a panel of mouse tissues. This revealed new expression signatures of known miRNAs as well as of novel miRNAs previously predicted using bioinformatic methods. Systematic confirmation of the array data with northern blotting and, in particular, real-time PCR suggests that the described microarray platform is a powerful tool to analyze miRNA expression patterns with rapid throughput and high fidelity.
Figures



References
-
- Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–4662. - PubMed
-
- Hammond SM. MicroRNAs as oncogenes. Curr. Opin. Genet. Dev. 2006;16:4–9. - PubMed
-
- Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309:1519–1524. - PubMed
-
- Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. - PubMed
-
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

