Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles
- PMID: 17356713
- PMCID: PMC1813932
- DOI: 10.1593/neo.07112
Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles
Abstract
DNA microarrays have been widely applied to cancer transcriptome analysis; however, the majority of such data are not easily accessible or comparable. Furthermore, several important analytic approaches have been applied to microarray analysis; however, their application is often limited. To overcome these limitations, we have developed Oncomine, a bioinformatics initiative aimed at collecting, standardizing, analyzing, and delivering cancer transcriptome data to the biomedical research community. Our analysis has identified the genes, pathways, and networks deregulated across 18,000 cancer gene expression microarrays, spanning the majority of cancer types and subtypes. Here, we provide an update on the initiative, describe the database and analysis modules, and highlight several notable observations. Results from this comprehensive analysis are available at http://www.oncomine.org.
Figures
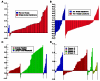

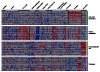
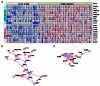
References
-
- Chung CH, Bernard PS, Perou CM. Molecular portraits and the family tree of cancer. Nat Genet. 2002;32:533–540. - PubMed
-
- Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. Large-scale metaanalysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. Proc Natl Acad Sci USA. 2004;101:9309–9314. (Epub 2004 June 9307) - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
