Patterns of host genome-wide gene transcript abundance in the peripheral blood of patients with acute dengue hemorrhagic fever
- PMID: 17357045
- PMCID: PMC4042601
- DOI: 10.1086/512162
Patterns of host genome-wide gene transcript abundance in the peripheral blood of patients with acute dengue hemorrhagic fever
Abstract
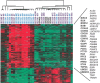
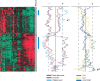
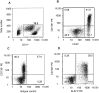
Responses by peripheral blood leukocytes may contribute to the pathogenesis of dengue hemorrhagic fever (DHF). We used DNA microarrays to reveal transcriptional patterns in the blood of 14 adults with DHF. Acute DHF was defined by an abundance of transcripts from cell cycle- and endoplasmic reticulum (ER)-related genes, suggesting a proliferative response accompanied by ER stress. Transcript-abundance levels for immunoresponse-associated genes, including cell surface markers, immunoglobulin, and innate response elements, were also elevated. Twenty-four genes were identified for which transcript abundance distinguished patients with dengue shock syndrome (DSS) from those without DSS. All the gene transcripts associated with DSS, many of which are induced by type I interferons, were less abundant in patients with DSS than in those without DSS. To our knowledge, these data provide the first snapshot of gene-expression patterns in peripheral blood during acute dengue and suggest that DSS is associated with attenuation of selected aspects of the innate host response.
Conflict of interest statement
Potential conflicts of interest: none reported.
Figures





References
-
- Libraty DH, Endy TP, Houng HS, et al. Differing influences of virus burden and immune activation on disease severity in secondary dengue-3 virus infections. J Infect Dis. 2002;185:1213–21. - PubMed
-
- Vaughn DW, Green S, Kalayanarooj S, et al. Dengue viremia titer, antibody response pattern, and virus serotype correlate with disease severity. J Infect Dis. 2000;181:2–9. - PubMed
-
- Green S, Vaughn DW, Kalayanarooj S, et al. Early immune activation in acute dengue illness is related to development of plasma leakage and disease severity. J Infect Dis. 1999;179:755–62. - PubMed
-
- Juffrie M, Meer GM, Hack CE, et al. Inflammatory mediators in dengue virus infection in children: interleukin-6 and its relation to C-reactive protein and secretory phospholipase A2. Am J Trop Med Hyg. 2001;65:70–5. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

