Overcoming the winner's curse: estimating penetrance parameters from case-control data
- PMID: 17357068
- PMCID: PMC1852705
- DOI: 10.1086/512821
Overcoming the winner's curse: estimating penetrance parameters from case-control data
Abstract
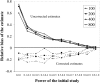
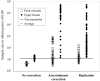
Genomewide association studies are now a widely used approach in the search for loci that affect complex traits. After detection of significant association, estimates of penetrance and allele-frequency parameters for the associated variant indicate the importance of that variant and facilitate the planning of replication studies. However, when these estimates are based on the original data used to detect the variant, the results are affected by an ascertainment bias known as the "winner's curse." The actual genetic effect is typically smaller than its estimate. This overestimation of the genetic effect may cause replication studies to fail because the necessary sample size is underestimated. Here, we present an approach that corrects for the ascertainment bias and generates an estimate of the frequency of a variant and its penetrance parameters. The method produces a point estimate and confidence region for the parameter estimates. We study the performance of this method using simulated data sets and show that it is possible to greatly reduce the bias in the parameter estimates, even when the original association study had low power. The uncertainty of the estimate decreases with increasing sample size, independent of the power of the original test for association. Finally, we show that application of the method to case-control data can improve the design of replication studies considerably.
Figures


References
Web Resource
-
- S.Z.'s Web site, http://www.sph.umich.edu/csg/zollner (for supplementary material containing results for additional genetic models, the size of the 95% confidence region for different sample sizes, and results for misspecified disease prevalence)
References
-
- Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K (2002) A comprehensive review of genetic association studies. Genet Med 4:45–61 - PubMed
-
- Capen EC, Clapp RV, Campbell WM (1971) Competitive bidding in high-risk situations. J Petrol Technol 23:641–653
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

