Characterization of Potocki-Lupski syndrome (dup(17)(p11.2p11.2)) and delineation of a dosage-sensitive critical interval that can convey an autism phenotype
- PMID: 17357070
- PMCID: PMC1852712
- DOI: 10.1086/512864
Characterization of Potocki-Lupski syndrome (dup(17)(p11.2p11.2)) and delineation of a dosage-sensitive critical interval that can convey an autism phenotype
Abstract
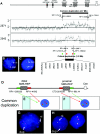
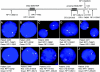
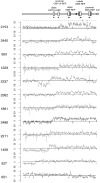
The duplication 17p11.2 syndrome, associated with dup(17)(p11.2p11.2), is a recently recognized syndrome of multiple congenital anomalies and mental retardation and is the first predicted reciprocal microduplication syndrome described--the homologous recombination reciprocal of the Smith-Magenis syndrome (SMS) microdeletion (del(17)(p11.2p11.2)). We previously described seven subjects with dup(17)(p11.2p11.2) and noted their relatively mild phenotype compared with that of individuals with SMS. Here, we molecularly analyzed 28 additional patients, using multiple independent assays, and also report the phenotypic characteristics obtained from extensive multidisciplinary clinical study of a subset of these patients. Whereas the majority of subjects (22 of 35) harbor the homologous recombination reciprocal product of the common SMS microdeletion (~3.7 Mb), 13 subjects (~37%) have nonrecurrent duplications ranging in size from 1.3 to 15.2 Mb. Molecular studies suggest potential mechanistic differences between nonrecurrent duplications and nonrecurrent genomic deletions. Clinical features observed in patients with the common dup(17)(p11.2p11.2) are distinct from those seen with SMS and include infantile hypotonia, failure to thrive, mental retardation, autistic features, sleep apnea, and structural cardiovascular anomalies. We narrow the critical region to a 1.3-Mb genomic interval that contains the dosage-sensitive RAI1 gene. Our results refine the critical region for Potocki-Lupski syndrome, provide information to assist in clinical diagnosis and management, and lend further support for the concept that genomic architecture incites genomic instability.
Figures




References
Web Resources
-
- Database of Genomic Variants, http://projects.tcag.ca/variation/ (for Human Genome Assembly build 36)
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for CMT1A, HNPP, SMS, RAI1, PMP22, and PLP1)
-
- Segmental Duplication Database, http://humanparalogy.gs.washington.edu/
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

