Characterizing an outbreak of vancomycin-resistant enterococci using hidden Markov models
- PMID: 17360254
- PMCID: PMC2373397
- DOI: 10.1098/rsif.2007.0224
Characterizing an outbreak of vancomycin-resistant enterococci using hidden Markov models
Abstract
Background: Antibiotic-resistant nosocomial pathogens can arise in epidemic clusters or sporadically. Genotyping is commonly used to distinguish epidemic from sporadic vancomycin-resistant enterococci (VRE). We compare this to a statistical method to determine the transmission characteristics of VRE.
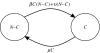
Methods and findings: A structured continuous-time hidden Markov model (HMM) was developed. The hidden states were the number of VRE-colonized patients (both detected and undetected). The input for this study was weekly point-prevalence data; 157 weeks of VRE prevalence. We estimated two parameters: one to quantify the cross-transmission of VRE and the other to quantify the level of VRE colonization from sporadic sources. We compared the results to those obtained by concomitant genotyping and phenotyping. We estimated that 89% of transmissions were due to ward cross-transmission while 11% were sporadic. Genotyping found that 90% had identical glycopeptide resistance genes and 84% were identical or nearly identical on pulsed-field gel electrophoresis (PFGE). There was some evidence, based on model selection criteria, that the cross-transmission parameter changed throughout the study period. The model that allowed for a change in transmission just prior to the outbreak and again at the peak of the outbreak was superior to other models. This model estimated that cross-transmission increased at week 120 and declined after week 135, coinciding with environmental decontamination.
Significance: We found that HMMs can be applied to serial prevalence data to estimate the characteristics of acquisition of nosocomial pathogens and distinguish between epidemic and sporadic acquisition. This model was able to estimate transmission parameters despite imperfect detection of the organism. The results of this model were validated against PFGE and glycopeptide resistance genotype data and produced very similar results. Additionally, HMMs can provide information about unobserved events such as undetected colonization.
Figures



Similar articles
-
Genotypic characterization of a nosocomial outbreak of VanA Enterococcus faecalis.Microb Drug Resist. 1996 Summer;2(2):231-7. doi: 10.1089/mdr.1996.2.231. Microb Drug Resist. 1996. PMID: 9158765
-
Mathematical modelling of vancomycin-resistant enterococci transmission during passive surveillance and active surveillance with contact isolation highlights the need to identify and address the source of acquisition.BMC Infect Dis. 2018 Oct 11;18(1):511. doi: 10.1186/s12879-018-3388-y. BMC Infect Dis. 2018. PMID: 30309313 Free PMC article.
-
Genotyping and preemptive isolation to control an outbreak of vancomycin-resistant Enterococcus faecium.Clin Infect Dis. 2006 Mar 15;42(6):739-46. doi: 10.1086/500322. Epub 2006 Feb 10. Clin Infect Dis. 2006. PMID: 16477546
-
[Vancomycin resistant enterococci in the Netherlands].Ned Tijdschr Geneeskd. 2004 May 1;148(18):878-82. Ned Tijdschr Geneeskd. 2004. PMID: 15152389 Review. Dutch.
-
Insights into the epidemiology and control of infection with vancomycin-resistant enterococci.Clin Infect Dis. 2000 Oct;31(4):1058-65. doi: 10.1086/318126. Epub 2000 Oct 25. Clin Infect Dis. 2000. PMID: 11049790 Review.
Cited by
-
A mathematical model and inference method for bacterial colonization in hospital units applied to active surveillance data for carbapenem-resistant enterobacteriaceae.PLoS One. 2020 Nov 12;15(11):e0231754. doi: 10.1371/journal.pone.0231754. eCollection 2020. PLoS One. 2020. PMID: 33180781 Free PMC article.
-
A Binary Prototype for Time-Series Surveillance and Intervention.medRxiv [Preprint]. 2025 Feb 5:2025.02.03.25321613. doi: 10.1101/2025.02.03.25321613. medRxiv. 2025. PMID: 39974019 Free PMC article. Preprint.
-
Quantifying where human acquisition of antibiotic resistance occurs: a mathematical modelling study.BMC Med. 2018 Aug 23;16(1):137. doi: 10.1186/s12916-018-1121-8. BMC Med. 2018. PMID: 30134939 Free PMC article.
-
Nosocomial transmission of C. difficile in English hospitals from patients with symptomatic infection.PLoS One. 2014 Jun 16;9(6):e99860. doi: 10.1371/journal.pone.0099860. eCollection 2014. PLoS One. 2014. PMID: 24932484 Free PMC article.
-
A modeling framework for the evolution and spread of antibiotic resistance: literature review and model categorization.Am J Epidemiol. 2013 Aug 15;178(4):508-20. doi: 10.1093/aje/kwt017. Epub 2013 May 9. Am J Epidemiol. 2013. PMID: 23660797 Free PMC article. Review.
References
-
- Bailey N. Charles Griffin; London, UK: 1975. The biomathematics of malaria.
-
- Bartley P.B, Schooneveldt J.M, Looke D.F, Morton A, Johnson D.W, Nimmo G.R. The relationship of a clonal outbreak of Enterococcus faecium VanA to methicillin-resistant Staphylococcus aureus incidence in an Australian hospital. J. Hosp. Infect. 2001;48:43–54. doi: 10.1053/jhin.2000.0915. - DOI - PubMed
-
- Baum L, Petrie T, Soules G, Weiss N. A maximisation technique occurring in the statistical analysis of probabilistic functions of Markov chains. Ann. Math. Stat. 1970;41:164–171.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

