The peptide pheromone-inducible conjugation system of Enterococcus faecalis plasmid pCF10: cell-cell signalling, gene transfer, complexity and evolution
- PMID: 17360276
- PMCID: PMC2435581
- DOI: 10.1098/rstb.2007.2043
The peptide pheromone-inducible conjugation system of Enterococcus faecalis plasmid pCF10: cell-cell signalling, gene transfer, complexity and evolution
Abstract
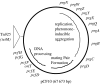
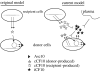
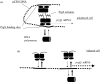
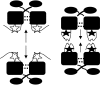
Expression of a large set of gene products required for conjugative transfer of the antibiotic resistance plasmid pCF10 is controlled by cell-cell communication between plasmid-free recipient cells and plasmid-carrying donor cells using a peptide mating pheromone cCF10. Most of the recent experimental analysis of this system has focused on the molecular events involved in initiation of the pheromone response in the donor cells, and on the mechanisms by which the donor cells control self-induction by endogenously produced pheromone. Recently, studies of the molecular machinery of conjugation encoded by the pheromone-inducible genes have been initiated. In addition, the system may serve as a useful bacterial model for addressing the evolution of biological complexity.
Figures
Similar articles
-
Identification and characterization of the genes of Enterococcus faecalis plasmid pCF10 involved in replication and in negative control of pheromone-inducible conjugation.Plasmid. 1996 Jan;35(1):46-57. doi: 10.1006/plas.1996.0005. Plasmid. 1996. PMID: 8693026
-
Antagonistic Donor Density Effect Conserved in Multiple Enterococcal Conjugative Plasmids.Appl Environ Microbiol. 2016 Jul 15;82(15):4537-45. doi: 10.1128/AEM.00363-16. Print 2016 Aug 1. Appl Environ Microbiol. 2016. PMID: 27208137 Free PMC article.
-
Molecular basis for control of conjugation by bacterial pheromone and inhibitor peptides.Mol Microbiol. 2006 Nov;62(4):958-69. doi: 10.1111/j.1365-2958.2006.05434.x. Epub 2006 Oct 13. Mol Microbiol. 2006. PMID: 17038121 Free PMC article.
-
Pheromone-inducible conjugation in Enterococcus faecalis: a model for the evolution of biological complexity?Int J Med Microbiol. 2006 Apr;296(2-3):141-7. doi: 10.1016/j.ijmm.2006.01.040. Epub 2006 Feb 28. Int J Med Microbiol. 2006. PMID: 16503196 Free PMC article. Review.
-
Enterococcal sex pheromones: signaling, social behavior, and evolution.Annu Rev Genet. 2013;47:457-82. doi: 10.1146/annurev-genet-111212-133449. Epub 2013 Sep 11. Annu Rev Genet. 2013. PMID: 24050179 Review.
Cited by
-
Genome Modification in Enterococcus faecalis OG1RF Assessed by Bisulfite Sequencing and Single-Molecule Real-Time Sequencing.J Bacteriol. 2015 Jun;197(11):1939-51. doi: 10.1128/JB.00130-15. Epub 2015 Mar 30. J Bacteriol. 2015. PMID: 25825433 Free PMC article.
-
Bioluminescence based biosensors for quantitative detection of enterococcal peptide-pheromone activity reveal inter-strain telesensing in vivo during polymicrobial systemic infection.Sci Rep. 2015 Feb 9;5:8339. doi: 10.1038/srep08339. Sci Rep. 2015. PMID: 25661457 Free PMC article.
-
Bacterial quorum sensing: its role in virulence and possibilities for its control.Cold Spring Harb Perspect Med. 2012 Nov 1;2(11):a012427. doi: 10.1101/cshperspect.a012427. Cold Spring Harb Perspect Med. 2012. PMID: 23125205 Free PMC article. Review.
-
RcgA and RcgR, Two Novel Proteins Involved in the Conjugative Transfer of Rhizobial Plasmids.mBio. 2022 Oct 26;13(5):e0194922. doi: 10.1128/mbio.01949-22. Epub 2022 Sep 8. mBio. 2022. PMID: 36073816 Free PMC article.
-
Biosynthesis of peptide signals in gram-positive bacteria.Adv Appl Microbiol. 2010;71:91-112. doi: 10.1016/S0065-2164(10)71004-2. Epub 2010 Feb 20. Adv Appl Microbiol. 2010. PMID: 20378052 Free PMC article. Review.
References
-
- Antiporta M.H, Dunny G.M. ccfA, the genetic determinant for the cCF10 peptide pheromone in Enterococcus faecalis OG1RF. J. Bacteriol. 2002;184:1155–1162. doi:10.1128/jb.184.4.1155-1162.2002 - DOI - PMC - PubMed
-
- Bae T, Dunny G.M. Dominant negative mutants of prgX: evidence for a role of PrgX dimerization in negative regulation of pheromone-inducible conjugation. Mol. Microbiol. 2001;39:1307–1320. doi:10.1111/j.1365-2958.2001.02319.x - DOI - PubMed
-
- Bae T, Clerc-Bardin S, Dunny G.M. Analysis of expression of prgX, a key negative regulator of the transfer of the Enterococcus faecalis pheromone-inducible plasmid pCF10. J. Mol. Biol. 2000;297:861–875. doi:10.1006/jmbi.2000.3628 - DOI - PubMed
-
- Bae T, Kozlowicz B, Dunny G.M. Two targets in pCF10 DNA for PrgX binding: their role in production of Qa and prgX mRNA and in regulation of pheromone-inducible conjugation. J. Mol. Biol. 2002;315:995–1007. doi:10.1006/jmbi.2001.5294 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

