Cell-cell communication in the plant pathogen Agrobacterium tumefaciens
- PMID: 17360279
- PMCID: PMC2435578
- DOI: 10.1098/rstb.2007.2040
Cell-cell communication in the plant pathogen Agrobacterium tumefaciens
Abstract
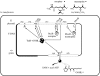
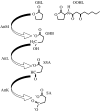
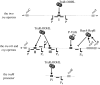
The plant pathogen Agrobacterium tumefaciens induces the formation of crown gall tumours at wound sites on host plants by directly transforming plant cells. This disease strategy benefits the bacteria as the infected plant tissue produces novel nutrients, called opines, that the colonizing bacteria can use as nutrients. Almost all of the genes that are required for virulence, and all of the opine uptake and utilization genes, are carried on large tumour-inducing (Ti) plasmids. The observation more than 25 years ago that specific opines are required for Ti plasmid conjugal transfer led to the discovery of a cell-cell signalling system on these plasmids that is similar to the LuxR-LuxI system first described in Vibrio fischeri. All Ti plasmids that have been described to date carry a functional LuxI-type N-acylhomoserine lactone synthase (TraI), and a LuxR-type signal receptor and transcriptional regulator called TraR. The traR genes are expressed only in the presence of specific opines called conjugal opines. The TraR-TraI system provides an important model for LuxR-LuxI-type systems, especially those found in the agriculturally important Rhizobiaceae family. In this review, we discuss current advances in the biochemistry and structural biology of the TraR-TraI system.
Figures

Similar articles
-
Co-evolution of the agrocinopine opines and the agrocinopine-mediated control of TraR, the quorum-sensing activator of the Ti plasmid conjugation system.Mol Microbiol. 2001 Sep;41(5):1173-85. doi: 10.1046/j.1365-2958.2001.02584.x. Mol Microbiol. 2001. PMID: 11555296
-
A LuxR-LuxI type regulatory system activates Agrobacterium Ti plasmid conjugal transfer in the presence of a plant tumor metabolite.J Bacteriol. 1994 May;176(10):2796-806. doi: 10.1128/jb.176.10.2796-2806.1994. J Bacteriol. 1994. PMID: 8188582 Free PMC article.
-
Quorum sensing but not autoinduction of Ti plasmid conjugal transfer requires control by the opine regulon and the antiactivator TraM.J Bacteriol. 2000 Feb;182(4):1080-8. doi: 10.1128/JB.182.4.1080-1088.2000. J Bacteriol. 2000. PMID: 10648535 Free PMC article.
-
Functions and regulation of quorum-sensing in Agrobacterium tumefaciens.Front Plant Sci. 2014 Jan 31;5:14. doi: 10.3389/fpls.2014.00014. eCollection 2014. Front Plant Sci. 2014. PMID: 24550924 Free PMC article. Review.
-
Agrobacterium tumefaciens: a Transformative Agent for Fundamental Insights into Host-Microbe Interactions, Genome Biology, Chemical Signaling, and Cell Biology.J Bacteriol. 2023 Apr 25;205(4):e0000523. doi: 10.1128/jb.00005-23. Epub 2023 Mar 9. J Bacteriol. 2023. PMID: 36892285 Free PMC article. Review.
Cited by
-
Coordination of division and development influences complex multicellular behavior in Agrobacterium tumefaciens.PLoS One. 2013;8(2):e56682. doi: 10.1371/journal.pone.0056682. Epub 2013 Feb 20. PLoS One. 2013. PMID: 23437210 Free PMC article.
-
D101 is critical for the function of AttJ, a repressor of quorum quenching system in Agrobacterium tumefaciens.J Microbiol. 2015 Sep;53(9):623-32. doi: 10.1007/s12275-015-5100-x. Epub 2015 Aug 1. J Microbiol. 2015. PMID: 26231372
-
The spent culture supernatant of Pseudomonas syringae contains azelaic acid.BMC Microbiol. 2018 Nov 28;18(1):199. doi: 10.1186/s12866-018-1352-z. BMC Microbiol. 2018. PMID: 30486794 Free PMC article.
-
Identification of an rsh gene from a Novosphingobium sp. necessary for quorum-sensing signal accumulation.J Bacteriol. 2009 Apr;191(8):2551-60. doi: 10.1128/JB.01692-08. Epub 2009 Feb 6. J Bacteriol. 2009. PMID: 19201802 Free PMC article.
-
Bacterial conversations: talking, listening and eavesdropping. An introduction.Philos Trans R Soc Lond B Biol Sci. 2007 Jul 29;362(1483):1115-7. doi: 10.1098/rstb.2007.2038. Philos Trans R Soc Lond B Biol Sci. 2007. PMID: 17360281 Free PMC article. Review. No abstract available.
References
-
- Beck von Bodman S, Hayman G.T, Farrand S.K. Opine catabolism and conjugal transfer of the nopaline Ti plasmid pTiC58 are coordinately regulated by a single repressor. Proc. Natl Acad. Sci. USA. 1992;89:643–647. doi:10.1073/pnas.89.2.643 - DOI - PMC - PubMed
-
- Binns A.N, Costantino P. The Agrobacterium oncogenes. In: Spaink H.P, Kondorosi A, Hooykaas P.J, editors. The Rhizobiaceae: molecular biology of model plant-associated bacteria. Kluwer Academic Publishers; Dordrecht, The Netherlands: 1998. pp. 251–266.
-
- Burr T.J, Otten L. Crown gall of grape: biology and disease management. Annu. Rev. Phytopathol. 1999;37:53–80. doi:10.1146/annurev.phyto.37.1.53 - DOI - PubMed
-
- Busby S, Ebright R.H. Transcription activation by catabolite activator protein (CAP) J. Mol. Biol. 1999;293:199–213. doi:10.1006/jmbi.1999.3161 - DOI - PubMed
-
- Carlier A, Uroz S, Smadja B, Fray R, Latour X, Dessaux Y, Faure D. The Ti plasmid of Agrobacterium tumefaciens harbors an attM-paralogous gene, aiiB, also encoding N-acyl homoserine lactonase activity. Appl. Environ. Microbiol. 2003;69:4989–4993. doi:10.1128/AEM.69.8.4989-4993.2003 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

