Mitotic recombination counteracts the benefits of genetic segregation
- PMID: 17360283
- PMCID: PMC2176173
- DOI: 10.1098/rspb.2007.0056
Mitotic recombination counteracts the benefits of genetic segregation
Abstract
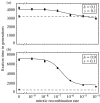
The ubiquity of sexual reproduction despite its cost has lead to an extensive body of research on the evolution and maintenance of sexual reproduction. Previous work has suggested that sexual reproduction can substantially speed up the rate of adaptation in diploid populations, because sexual populations are able to produce the fittest homozygous genotype by segregation and mating of heterozygous individuals. In contrast, asexual populations must wait for two rare mutational events, one producing a heterozygous carrier and the second converting a heterozygous to a homozygous carrier, before a beneficial mutation can become fixed. By avoiding this additional waiting time, it was shown that the benefits of segregation could overcome a twofold cost of sex. This previous result ignores mitotic recombination (MR), however. Here, we show that MR significantly hastens the spread of beneficial mutations in asexual populations. Indeed, given empirical data on MR, we find that adaptation in asexual populations proceeds as fast as that in sexual populations, especially when beneficial alleles are partially recessive. We conclude that asexual populations can gain most of the benefit of segregation through MR while avoiding the costs associated with sexual reproduction.
Figures






References
-
- Agrawal A.F, Otto S.P. Host–parasite coevolution and selection on sex through the effects of segregation. Am. Nat. 2006;168:617–629. doi:10.1086/508029 - DOI - PubMed
-
- Aguilera A, Chavez S, Malagon F. Mitotic recombination in yeast: elements controlling its incidence. Yeast. 2000;16:731–754. doi:10.1002/1097-0061(20000615)16:8<731::AID-YEA586>3.0.CO;2-L - DOI - PubMed
-
- Antezana M, Hudson R.R. Before crossing over: the advantages of eukaryotic sex in genomes lacking chiasmatic recombination. Genet. Res. 1997a;70:7–25. doi:10.1017/S0016672397002875 - DOI - PubMed
-
- Antezana M, Hudson R.R. Era reversibile! Point mutations, the ratchet, and the initial success of eukaryotic sex: a simulation study. Evol. Theory. 1997b;11:209–235.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

