Sensitive ChIP-DSL technology reveals an extensive estrogen receptor alpha-binding program on human gene promoters
- PMID: 17360330
- PMCID: PMC1821125
- DOI: 10.1073/pnas.0700715104
Sensitive ChIP-DSL technology reveals an extensive estrogen receptor alpha-binding program on human gene promoters
Abstract
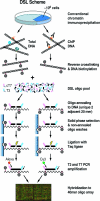
ChIP coupled with microarray provides a powerful tool to determine in vivo binding profiling of transcription factors to deduce regulatory circuitries in mammalian cells. Aiming at improving the specificity and sensitivity of such analysis, we developed a new technology called ChIP-DSL using the DNA selection and ligation (DSL) strategy, permitting robust analysis with much reduced materials compared with standard procedures. We profiled general and sequence-specific DNA binding transcription factors using a full human genome promoter array based on the ChIP-DSL technology, revealing an unprecedented number of the estrogen receptor (ERalpha) target genes in MCF-7 cells. Coupled with gene expression profiling, we found that only a fraction of these direct ERalpha target genes were highly responsive to estrogen and that the expression of those ERalpha-bound, estrogen-inducible genes was associated with breast cancer progression in humans. This study demonstrates the power of the ChIP-DSL technology in revealing regulatory gene expression programs that have been previously invisible in the human genome.
Figures




References
-
- Strahl BD, Allis CD. Nature. 2000;403:41–45. - PubMed
-
- Fischle W, Wang Y, Allis CD. Curr Opin Cell Biol. 2003;15:172–183. - PubMed
-
- Turner BM. BioEssays. 2000;22:836–845. - PubMed
-
- Sims RJ, III, Nishioka K, Reinberg D. Trends Genet. 2003;19:629–639. - PubMed
-
- Rosenfeld MG, Lunyak VV, Glass CK. Genes Dev. 2006;20:1405–1428. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

