An epigenetic mark generated by the incorporation of CENP-A into centromeric nucleosomes
- PMID: 17360341
- PMCID: PMC1829255
- DOI: 10.1073/pnas.0700390104
An epigenetic mark generated by the incorporation of CENP-A into centromeric nucleosomes
Abstract
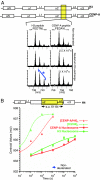
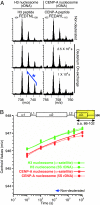
Mammalian centromeres are defined epigenetically. Although the physical nature of the epigenetic mark is unknown, nucleosomes in which CENP-A replaces histone H3 are at the foundation of centromeric chromatin. Hydrogen/deuterium exchange MS is now used to show that assembly into nucleosomes imposes stringent conformational constraints, reducing solvent accessibility in almost all histone regions by >3 orders of magnitude. Despite this, nucleosomes assembled with CENP-A are substantially more conformationally rigid than those assembled with histone H3 independent of DNA template. Substitution of the CENP-A centromere targeting domain into histone H3 to convert it into a centromere-targeted histone that can functionally replace CENP-A in centromere maintenance generates the same more rigid nucleosome, as does CENP-A. Thus, the targeting information directing CENP-A deposition at the centromere produces a structurally distinct nucleosome, supporting a CENP-A-driven self-assembly mechanism that mediates maintenance of centromere identity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Strahl BD, Allis CD. Nature. 2000;403:41–45. - PubMed
-
- Cleveland DW, Mao Y, Sullivan KF. Cell. 2003;112:407–421. - PubMed
-
- Carroll CW, Straight AF. Trends Cell Biol. 2006;16:70–78. - PubMed
-
- Jiang J, Birchler JA, Parrott WA, Dawe RK. Trends Plant Sci. 2003;8:570–575. - PubMed
-
- Willard HF, Waye JS. Trends Genet. 1987;3:192–198.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

