A lamin A protein isoform overexpressed in Hutchinson-Gilford progeria syndrome interferes with mitosis in progeria and normal cells
- PMID: 17360355
- PMCID: PMC1821129
- DOI: 10.1073/pnas.0611640104
A lamin A protein isoform overexpressed in Hutchinson-Gilford progeria syndrome interferes with mitosis in progeria and normal cells
Abstract
Hutchinson-Gilford progeria syndrome (HGPS) is a rare genetic disorder characterized by dramatic premature aging. Classic HGPS is caused by a de novo point mutation in exon 11 (residue 1824, C --> T) of the LMNA gene, activating a cryptic splice donor and resulting in a mutant lamin A (LA) protein termed "progerin/LADelta50" that lacks the normal cleavage site to remove a C-terminal farnesyl group. During interphase, irreversibly farnesylated progerin/LADelta50 anchors to the nuclear membrane and causes characteristic nuclear blebbing. Progerin/LADelta50's localization and behavior during mitosis, however, are completely unknown. Here, we report that progerin/LADelta50 mislocalizes into insoluble cytoplasmic aggregates and membranes during mitosis and causes abnormal chromosome segregation and binucleation. These phenotypes are largely rescued with either farnesyltransferase inhibitors or a farnesylation-incompetent mutant progerin/LADelta50. Furthermore, we demonstrate that small amounts of progerin/LADelta50 exist in normal fibroblasts, and a significant percentage of these progerin/LADelta50-expressing normal cells are binucleated, implicating progerin/LADelta50 as causing similar mitotic defects in the normal aging process. Our findings present evidence of mitotic abnormality in HGPS and may shed light on the general phenomenon of aging.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
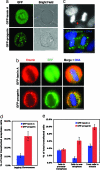
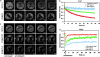

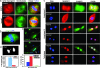

References
-
- Gruenbaum Y, Margalit A, Goldman RD, Shumaker DK, Wilson KL. Nat Rev Mol Cell Biol. 2005;6:21–31. - PubMed
-
- Goldman RD, Gruenbaum Y, Moir RD, Shumaker DK, Spann TP. Genes Dev. 2002;16:533–547. - PubMed
-
- Sinensky M, Fantle K, Trujillo M, McLain T, Kupfer A, Dalton M. J Cell Sci. 1994;107:61–67. - PubMed
-
- Capell B, Collins F. Nat Rev Genet. 2006;7:940–952. - PubMed
-
- DeBusk FL. J Pediatr. 1972;80:697–724. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

