RNA aptamers directed to discrete functional sites on a single protein structural domain
- PMID: 17360423
- PMCID: PMC1820654
- DOI: 10.1073/pnas.0607805104
RNA aptamers directed to discrete functional sites on a single protein structural domain
Abstract
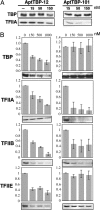
Cellular regulatory networks are organized such that many proteins have few interactions, whereas a few proteins have many. These densely connected protein "hubs" are critical for the system-wide behavior of cells, and the capability of selectively perturbing a subset of interactions at these hubs is invaluable in deciphering and manipulating regulatory mechanisms. SELEX-generated RNA aptamers are proving to be highly effective reagents for inhibiting targeted proteins, but conventional methods generate one or several aptamer clones that usually bind to a single target site most preferred by a nucleic acid ligand. We advance a generalized scheme for isolating aptamers to multiple sites on a target molecule by reducing the ability of the preferred site to select its cognate aptamer. We demonstrate the use of this scheme by generating aptamers directed to discrete functional surfaces of the yeast TATA-binding protein (TBP). Previously we selected "class 1" RNA aptamers that interfere with the TBP's binding to TATA-DNA. By masking TBP with TATA-DNA or an unamplifiable class 1 aptamer, we isolated a new aptamer class, "class 2," that can bind a TBP.DNA complex and is in competition with binding another general transcription factor, TFIIA. Moreover, we show that both of these aptamers inhibit RNA polymerase II-dependent transcription, but analysis of template-bound factors shows they do so in mechanistically distinct and unexpected ways that can be attributed to binding either the DNA or TFIIA recognition sites. These results should spur innovative approaches to modulating other highly connected regulatory proteins.
Conflict of interest statement
Conflict of interest statement: H.S. and J.T.L. have a patent and patent submissions on RNA aptamers and aptamer-related methods.
Figures


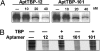
Similar articles
-
Perturbation of discrete sites on a single protein domain with RNA aptamers: targeting of different sides of the TATA-binding protein (TBP).Biosci Biotechnol Biochem. 2013;77(8):1739-46. doi: 10.1271/bbb.130296. Epub 2013 Aug 7. Biosci Biotechnol Biochem. 2013. PMID: 23924740
-
Novel interactions between the components of human and yeast TFIIA/TBP/DNA complexes.J Mol Biol. 2003 Sep 26;332(4):783-93. doi: 10.1016/s0022-2836(03)00887-8. J Mol Biol. 2003. PMID: 12972251
-
A sol-gel-based microfluidics system enhances the efficiency of RNA aptamer selection.Oligonucleotides. 2011 Mar-Apr;21(2):93-100. doi: 10.1089/oli.2010.0263. Epub 2011 Mar 17. Oligonucleotides. 2011. PMID: 21413890
-
Role of the TATA-box binding protein (TBP) and associated family members in transcription regulation.Gene. 2022 Jul 30;833:146581. doi: 10.1016/j.gene.2022.146581. Epub 2022 May 18. Gene. 2022. PMID: 35597524 Review.
-
X-ray crystallographic studies of eukaryotic transcription initiation factors.Philos Trans R Soc Lond B Biol Sci. 1996 Apr 29;351(1339):483-9. doi: 10.1098/rstb.1996.0046. Philos Trans R Soc Lond B Biol Sci. 1996. PMID: 8735270 Review.
Cited by
-
Fluorescence-Based Strategies to Investigate the Structure and Dynamics of Aptamer-Ligand Complexes.Front Chem. 2016 Aug 3;4:33. doi: 10.3389/fchem.2016.00033. eCollection 2016. Front Chem. 2016. PMID: 27536656 Free PMC article. Review.
-
Identification of Sequence-Selective Tyrosine Kinase Deoxyribozymes.J Mol Evol. 2015 Dec;81(5-6):218-24. doi: 10.1007/s00239-015-9699-3. Epub 2015 Sep 25. J Mol Evol. 2015. PMID: 26407964 Free PMC article.
-
Development of single-stranded DNA aptamers for specific Bisphenol a detection.Oligonucleotides. 2011 Mar-Apr;21(2):85-91. doi: 10.1089/oli.2010.0267. Epub 2011 Mar 17. Oligonucleotides. 2011. PMID: 21413891 Free PMC article.
-
Nucleic acid aptamers for targeting of shRNA-based cancer therapeutics.Biologics. 2007 Dec;1(4):367-76. Biologics. 2007. PMID: 19707307 Free PMC article.
-
Delivery systems for in vivo use of nucleic Acid drugs.Drug Target Insights. 2007;2:183-96. Epub 2007 Aug 9. Drug Target Insights. 2007. PMID: 21901073 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

