Mechanisms controlling the acquisition of a cardiac phenotype by liver stem cells
- PMID: 17360446
- PMCID: PMC1805456
- DOI: 10.1073/pnas.0700416104
Mechanisms controlling the acquisition of a cardiac phenotype by liver stem cells
Abstract
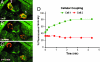
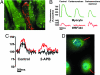
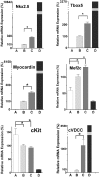
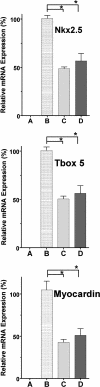
The mechanisms underlying stem cell acquisition of a cardiac phenotype are unresolved. We studied early events during the acquisition of a cardiac phenotype by a cloned adult liver stem cell line (WB F344) in a cardiac microenvironment. WB F344 cells express a priori the transcription factors GATA4 and SRF, connexin 43 in the cell membrane, and myoinositol 1,4,5-triphosphate receptor in the perinuclear region. Functional cell-cell communication developed between WB F344 cells and adjacent cocultured cardiomyocytes in 24 h. De novo cytoplasmic [Ca(2+)](c) and nuclear [Ca(2+)](nu) oscillations appeared in WB F344 cells, synchronous with [Ca(2+)](i) transients in adjacent cardiomyocytes. The [Ca(2+)] oscillations in the WB F344 cells, but not those in the cardiomyocytes, were eliminated by a gap junction uncoupler and reappeared with its removal. By 24 h, WB F344 cells began expressing the cardiac transcription factors Nkx2.5, Tbx5, and cofactor myocardin; cardiac proteins 24 h later; and a sarcomeric pattern 4-6 days later. Myoinositol 1,4,5-triphosphate receptor inhibition suppressed WB F344 cell [Ca(2+)](nu) oscillations but not [Ca(2+)](c) oscillations, and L-type calcium channel inhibition eliminated [Ca(2+)] oscillations in cardiomyocytes and WB F344 cells. The use of these inhibitors was associated with a decrease in Nkx2.5, Tbx5, and myocardin expression in the WB F344 cells. Our findings suggest that signals from cardiomyocytes diffuse through shared channels, inducing [Ca(2+)] oscillations in the WB F344 cells. We hypothesize that the WB F344 cell [Ca(2+)](nu) oscillations activate the expression of a cardiac specifying gene program, ushering in a cardiac phenotype.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Anversa P, Leri A, Kajstura J. J Am Coll Cardiol. 2006;47:1769–1776. - PubMed
-
- Murry CE, Reinecke H, Pabon LM. J Am Coll Cardiol. 2006;47:1777–1785. - PubMed
-
- Alvarez-Dolado M, Pardal R, Garcia-Verdugo JM, Fike JR, Lee HO, Pfeffer K, Lois C, Morrison SJ, Alvarez-Buylla A. Nature. 2003;425:968–973. - PubMed
-
- Kajstura J, Rota M, Whang B, Cascapera S, Hosoda T, Bearzi C, Nurzynska D, Kasahara H, Zias E, Bonafe M, et al. Circ Res. 2005;96:127–137. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

