Adaptive evolution in humans revealed by the negative correlation between the polymorphism and fixation phases of evolution
- PMID: 17360451
- PMCID: PMC1820682
- DOI: 10.1073/pnas.0605565104
Adaptive evolution in humans revealed by the negative correlation between the polymorphism and fixation phases of evolution
Abstract
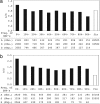
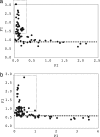
The selective forces acting on amino acid substitutions may be different in the two phases of molecular evolution: polymorphism and fixation. Negative selection and genetic drift may dominate the first phase, whereas positive selection may become much more significant in the second phase. However, the conventional dichotomy of synonymous vs. nonsynonymous changes does not offer the resolution needed to study the dynamics of these two phases. Following previously published methods, we separated amino acid changes into 75 elementary types (1-bp substitution between their respective codons). The likelihood of each type of amino acid change becoming polymorphic (PI, which stands for "polymorphic index"), relative to synonymous changes, can then be calculated. Similarly, the likelihood of fixation (FI, for "fixation index"), conditional on common polymorphisms, is also calculated. Using Perlegen and HapMap data on human polymorphisms and the chimpanzee sequences as the outgroup, we compared the evolutionary dynamics of the 75 elementary changes in the two phases. We found a strong "L-shaped" negative correlation (P < 0.001) between FI and PI. Only those changes with low PIs show FI > 1, which is often a signature of adaptive evolution. These patterns suggest that negative and positive selection operate more effectively on the same set of amino acid changes and that approximately 10-13% of amino acid substitutions between humans and chimpanzee may be adaptive.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

