Pollen tetrad-based visual assay for meiotic recombination in Arabidopsis
- PMID: 17360452
- PMCID: PMC1805420
- DOI: 10.1073/pnas.0608936104
Pollen tetrad-based visual assay for meiotic recombination in Arabidopsis
Abstract
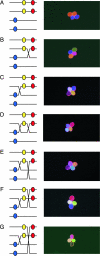
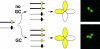
Recombination, in the form of cross-overs (COs) and gene conversion (GC), is a highly conserved feature of meiosis from fungi to mammals. Recombination helps ensure chromosome segregation and promotes allelic diversity. Lesions in the recombination machinery are often catastrophic for meiosis, resulting in sterility. We have developed a visual assay capable of detecting Cos and GCs and measuring CO interference in Arabidopsis thaliana. This flexible assay utilizes transgene constructs encoding pollen-expressed fluorescent proteins of three different colors in the qrt1 mutant background. By observing the segregation of the fluorescent alleles in 92,489 pollen tetrads, we demonstrate (i) a correlation between developmental position and CO frequency, (ii) a temperature dependence for CO frequency, (iii) the ability to detect meiotic GC events, and (iv) the ability to rapidly assess CO interference.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Comment in
-
Meiosis in living color: fluorescence-based tetrad analysis in Arabidopsis.Proc Natl Acad Sci U S A. 2007 Mar 6;104(10):3673-4. doi: 10.1073/pnas.0700276104. Epub 2007 Feb 28. Proc Natl Acad Sci U S A. 2007. PMID: 17360410 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

