DgrA is a member of a new family of cyclic diguanosine monophosphate receptors and controls flagellar motor function in Caulobacter crescentus
- PMID: 17360486
- PMCID: PMC1805490
- DOI: 10.1073/pnas.0607738104
DgrA is a member of a new family of cyclic diguanosine monophosphate receptors and controls flagellar motor function in Caulobacter crescentus
Erratum in
- Proc Natl Acad Sci U S A. 2007 May 1;104(18):7729
Abstract
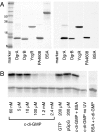
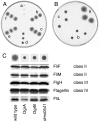
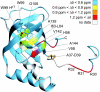
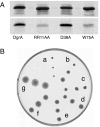
Bacteria are able to switch between two mutually exclusive lifestyles, motile single cells and sedentary multicellular communities that colonize surfaces. These behavioral changes contribute to an increased fitness in structured environments and are controlled by the ubiquitous bacterial second messenger cyclic diguanosine monophosphate (c-di-GMP). In response to changing environments, fluctuating levels of c-di-GMP inversely regulate cell motility and cell surface adhesins. Although the synthesis and breakdown of c-di-GMP has been studied in detail, little is known about the downstream effector mechanisms. Using affinity chromatography, we have isolated several c-di-GMP-binding proteins from Caulobacter crescentus. One of these proteins, DgrA, is a PilZ homolog involved in mediating c-di-GMP-dependent control of C. crescentus cell motility. Biochemical and structural analysis of DgrA and homologs from C. crescentus, Salmonella typhimurium, and Pseudomonas aeruginosa demonstrated that this protein family represents a class of specific diguanylate receptors and suggested a general mechanism for c-di-GMP binding and signal transduction. Increased concentrations of c-di-GMP or DgrA blocked motility in C. crescentus by interfering with motor function rather than flagellar assembly. We present preliminary evidence implicating the flagellar motor protein FliL in DgrA-dependent cell motility control.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

