HIV-1 Vpr function is mediated by interaction with the damage-specific DNA-binding protein DDB1
- PMID: 17360488
- PMCID: PMC1820720
- DOI: 10.1073/pnas.0610167104
HIV-1 Vpr function is mediated by interaction with the damage-specific DNA-binding protein DDB1
Abstract
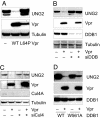
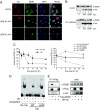
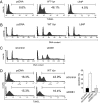
The Vpr accessory protein of HIV-1 induces a response similar to that of DNA damage. In cells expressing Vpr, the DNA damage sensing kinase, ATR, is activated, resulting in G(2) arrest and apoptosis. In addition, Vpr causes rapid degradation of the uracil-DNA glycosylases UNG2 and SMUG1. Although several cellular proteins have been reported to bind to Vpr, the mechanism by which Vpr mediates its biological effects is unknown. Using tandem affinity purification and mass spectrometry, we identified a predominant cellular protein that binds to Vpr as the damage-specific DNA-binding protein 1 (DDB1). In addition to its role in the repair of damaged DNA, DDB1 is a component of an E3 ubiquitin ligase that degrades numerous cellular substrates. Interestingly, DDB1 is targeted by specific regulatory proteins of other viruses, including simian virus 5 and hepatitis B. We show that the interaction with DDB1 mediates Vpr-induced apoptosis and UNG2/SMUG1 degradation and impairs the repair of UV-damaged DNA, which could account for G(2) arrest and apoptosis. The interaction with DDB1 may explain several of the diverse biological functions of Vpr and suggests potential roles for Vpr in HIV-1 replication.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Sherman MP, De Noronha CM, Williams SA, Greene WC. DNA Cell Biol. 2002;21:679–688. - PubMed
-
- Muthumani K, Desai BM, Hwang DS, Choo AY, Laddy DJ, Thieu KP, Rao RG, Weiner DB. DNA Cell Biol. 2004;23:239–247. - PubMed
-
- Goh WC, Rogel ME, Kinsey CM, Michael SF, Fultz PN, Nowak MA, Hahn BH, Emerman M. Nat Med. 1998;4:65–71. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

