EDEM1 reveals a quality control vesicular transport pathway out of the endoplasmic reticulum not involving the COPII exit sites
- PMID: 17360537
- PMCID: PMC1810509
- DOI: 10.1073/pnas.0700154104
EDEM1 reveals a quality control vesicular transport pathway out of the endoplasmic reticulum not involving the COPII exit sites
Abstract
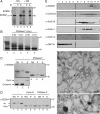
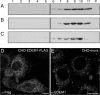
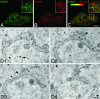
Immature and nonnative proteins are retained in the endoplasmic reticulum (ER) by the quality control machinery. Folding-incompetent glycoproteins are eventually targeted for ER-associated protein degradation (ERAD). EDEM1 (ER degradation-enhancing alpha-mannosidase-like protein 1), a putative mannose-binding protein, targets misfolded glycoproteins for ERAD. We report that endogenous EDEM1 exists mainly as a soluble glycoprotein. By high-resolution immunolabeling and serial section analysis, we find that endogenous EDEM1 is sequestered in buds that form along cisternae of the rough ER at regions outside of the transitional ER. They give rise to approximately 150-nm vesicles scattered throughout the cytoplasm that are lacking a recognizable COPII coat. About 87% of the immunogold labeling was over the vesicles and approximately 11% over the ER lumen. Some of the EDEM1 vesicles also contain Derlin-2 and the misfolded Hong Kong variant of alpha-1-antitrypsin, a substrate for EDEM1 and ERAD. Our results demonstrate the existence of a vesicle budding transport pathway out of the rough ER that does not involve the canonical transitional ER exit sites and therefore represents a previously unrecognized passageway to remove potentially harmful misfolded luminal glycoproteins from the ER.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
EDEM1 recognition and delivery of misfolded proteins to the SEL1L-containing ERAD complex.Mol Cell. 2009 Jun 12;34(5):627-33. doi: 10.1016/j.molcel.2009.05.018. Mol Cell. 2009. PMID: 19524542 Free PMC article.
-
EDEM1 regulates ER-associated degradation by accelerating de-mannosylation of folding-defective polypeptides and by inhibiting their covalent aggregation.Biochem Biophys Res Commun. 2006 Nov 3;349(4):1278-84. doi: 10.1016/j.bbrc.2006.08.186. Epub 2006 Sep 12. Biochem Biophys Res Commun. 2006. PMID: 16987498
-
Mannose trimming is required for delivery of a glycoprotein from EDEM1 to XTP3-B and to late endoplasmic reticulum-associated degradation steps.J Biol Chem. 2011 Jan 14;286(2):1292-300. doi: 10.1074/jbc.M110.154849. Epub 2010 Nov 9. J Biol Chem. 2011. PMID: 21062743 Free PMC article.
-
Quality control of glycoprotein folding and ERAD: the role of N-glycan handling, EDEM1 and OS-9.Histochem Cell Biol. 2017 Feb;147(2):269-284. doi: 10.1007/s00418-016-1513-9. Epub 2016 Nov 1. Histochem Cell Biol. 2017. PMID: 27803995 Review.
-
Mechanisms of COPII vesicle formation and protein sorting.FEBS Lett. 2007 May 22;581(11):2076-82. doi: 10.1016/j.febslet.2007.01.091. Epub 2007 Feb 14. FEBS Lett. 2007. PMID: 17316621 Review.
Cited by
-
Endoplasmic reticulum (ER) mannosidase I is compartmentalized and required for N-glycan trimming to Man5-6GlcNAc2 in glycoprotein ER-associated degradation.Mol Biol Cell. 2008 Jan;19(1):216-25. doi: 10.1091/mbc.e07-05-0505. Epub 2007 Nov 14. Mol Biol Cell. 2008. PMID: 18003979 Free PMC article.
-
Glycoprotein folding and quality-control mechanisms in protein-folding diseases.Dis Model Mech. 2014 Mar;7(3):331-41. doi: 10.1242/dmm.014589. Dis Model Mech. 2014. PMID: 24609034 Free PMC article. Review.
-
Flagging and docking: dual roles for N-glycans in protein quality control and cellular proteostasis.Trends Biochem Sci. 2012 Oct;37(10):404-10. doi: 10.1016/j.tibs.2012.07.005. Epub 2012 Aug 23. Trends Biochem Sci. 2012. PMID: 22921611 Free PMC article.
-
Coronaviruses construct an interconnection way with ERAD and autophagy.Future Microbiol. 2021 Sep;16(14):1135-1151. doi: 10.2217/fmb-2021-0044. Epub 2021 Sep 1. Future Microbiol. 2021. PMID: 34468179 Free PMC article. Review.
-
Selective autophagy of cytosolic protein aggregates involves ribosome-free rough endoplasmic reticulum.Histochem Cell Biol. 2020 Feb;153(2):89-99. doi: 10.1007/s00418-019-01829-w. Epub 2019 Nov 12. Histochem Cell Biol. 2020. PMID: 31720797
References
-
- Ellgaard L, Helenius A. Nat Rev Mol Cell Biol. 2003;4:181–191. - PubMed
-
- Petrucelli L, Dawson TM. Ann Med. 2004;36:315–320. - PubMed
-
- Aridor M, Hannan LA. Traffic. 2002;3:781–790. - PubMed
-
- Molinari M, Helenius A. Science. 2000;288:331–333. - PubMed
-
- Trombetta ES, Parodi AJ. Annu Rev Cell Dev Biol. 2003;19:649–676. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

