A statistical phylogeography of influenza A H5N1
- PMID: 17360548
- PMCID: PMC1838625
- DOI: 10.1073/pnas.0700435104
A statistical phylogeography of influenza A H5N1
Abstract
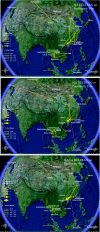
The geographic diffusion of highly pathogenic influenza A H5N1 has largely been traced from the perspective of the virus's victims. Birds of a variety of avian orders have been sampled across localities, and their infection has been identified by a general genetic test. Another approach tracks the migration from the perspective of the virus alone, by way of a phylogeography of H5N1 genetic sequences. Although several phylogenies in the literature have labeled H5N1 clades by geographic region, none has analytically inferred the history of the virus's migration. With a statistical phylogeography of 192 hemagglutinin and neuraminidase isolates, we show that the Chinese province of Guangdong is the source of multiple H5N1 strains spreading at both regional and international scales. In contrast, Indochina appears to be a regional sink, at the same time demonstrating bidirectional dispersal among localities within the region. An evolutionary trace of HA(1) across the phylogeography suggests a mechanism by which H5N1 is able to infect repeated cycles of host species across localities, regardless of the host species first infected in each locale. The trace also hypothesizes amino acid replacements that preceded the first recorded outbreak of pathogenic H5N1 in Hong Kong, 1997.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

