Haplotype structure and selection of the MDM2 oncogene in humans
- PMID: 17360557
- PMCID: PMC1838634
- DOI: 10.1073/pnas.0610998104
Haplotype structure and selection of the MDM2 oncogene in humans
Abstract
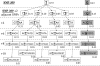
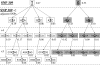
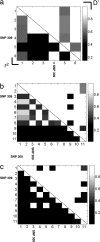
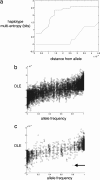
The MDM2 protein is an ubiquitin ligase that plays a critical role in regulating the levels and activity of the p53 protein, which is a central tumor suppressor. A SNP in the human MDM2 gene (SNP309 T/G) occurs at frequencies dependent on demographic history and has been shown to have important differential effects on the activity of the MDM2 and p53 proteins and to associate with altered risk for the development of several cancers. In this report, the haplotype structure of the MDM2 gene is determined by using 14 different SNPs across the gene from three different population samples: Caucasians, African Americans, and the Ashkenazi Jewish ethnic group. The results presented in this report indicate that there is a substantially reduced variability of the deleterious SNP309 G allele haplotype in all three populations studied, whereas multiple common T allele haplotypes were found in all three populations. This observation, coupled with the relatively high frequency of the G allele haplotype in both and Caucasian and Ashkenazi Jewish population data sets, suggests that this haplotype could have undergone a recent positive selection sweep. An entropy-based selection test is presented that explicitly takes into account the correlations between different SNPs, and the analysis of MDM2 reveals a significant departure from the standard assumptions of selective neutrality.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Jin S, Levine AJ. J Cell Sci. 2001;114:4139–4140. - PubMed
-
- Lain S, Lane D. Eur J Cancer. 2003;39:1053–1060. - PubMed
-
- Bond GL, Hu W, Levine AJ. Curr Cancer Drug Targets. 2005;5:3–8. - PubMed
-
- Murphy ME. Cell Death Differ. 2006;13:916–920. - PubMed
-
- Bonafe M, Salvioli S, Barbi C, Mishto M, Trapassi C, Gemelli C, Storci G, Olivieri F, Monti D, Franceschi C. Biochem Biophys Res Commun. 2002;299:539–541. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

