In vitro analysis of DNA-protein interactions by proximity ligation
- PMID: 17360610
- PMCID: PMC1805562
- DOI: 10.1073/pnas.0611229104
In vitro analysis of DNA-protein interactions by proximity ligation
Abstract
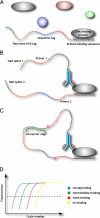
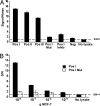
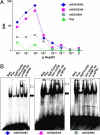
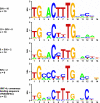
Protein-binding DNA sequence elements encode a variety of regulated functions of genomes. Information about such elements is currently in a state of rapid growth, but improved methods are required to characterize the sequence specificity of DNA-binding proteins. We have established an in vitro method for specific and sensitive solution-phase analysis of interactions between proteins and nucleic acids in nuclear extracts, based on the proximity ligation assay. The reagent consumption is very low, and the excellent sensitivity of the assay enables analysis of as few as 1-10 cells. We show that our results are highly reproducible, quantitative, and in good agreement with both EMSA and predictions obtained by using a motif finding software. This assay can be a valuable tool to characterize in-depth the sequence specificity of DNA-binding proteins and to evaluate effects of polymorphisms in known transcription factor binding sites.
Conflict of interest statement
Conflict of interest statement: U.L. is the inventor of patents describing the proximity ligation technology. He is one of the founders of the company Olink AB, which exploits the proximity ligation technology.
Figures
Similar articles
-
DNA-protein interactions: methods for detection and analysis.Mol Cell Biochem. 2012 Jun;365(1-2):279-99. doi: 10.1007/s11010-012-1269-z. Epub 2012 Mar 8. Mol Cell Biochem. 2012. PMID: 22399265 Review.
-
Some microsatellites may act as novel polymorphic cis-regulatory elements through transcription factor binding.Gene. 2004 Oct 27;341:149-65. doi: 10.1016/j.gene.2004.06.035. Gene. 2004. PMID: 15474298
-
DNA polymorphisms in potential regulatory elements of the CFTR gene alter transcription factor binding.Hum Genet. 2002 Jul;111(1):66-74. doi: 10.1007/s00439-002-0737-z. Epub 2002 Jun 6. Hum Genet. 2002. PMID: 12136238
-
Visualising individual sequence-specific protein-DNA interactions in situ.N Biotechnol. 2012 Jun 15;29(5):589-98. doi: 10.1016/j.nbt.2011.08.002. Epub 2011 Aug 31. N Biotechnol. 2012. PMID: 21906700
-
Proximity ligation assay: an ultrasensitive method for protein quantification and its applications in pathogen detection.Appl Microbiol Biotechnol. 2021 Feb;105(3):923-935. doi: 10.1007/s00253-020-11049-1. Epub 2021 Jan 11. Appl Microbiol Biotechnol. 2021. PMID: 33427935 Review.
Cited by
-
DNA-protein interactions: methods for detection and analysis.Mol Cell Biochem. 2012 Jun;365(1-2):279-99. doi: 10.1007/s11010-012-1269-z. Epub 2012 Mar 8. Mol Cell Biochem. 2012. PMID: 22399265 Review.
-
Interferon regulatory factor 5 (IRF5) gene variants are associated with multiple sclerosis in three distinct populations.J Med Genet. 2008 Jun;45(6):362-9. doi: 10.1136/jmg.2007.055012. Epub 2008 Feb 19. J Med Genet. 2008. PMID: 18285424 Free PMC article.
-
Sensitive plasma protein analysis by microparticle-based proximity ligation assays.Mol Cell Proteomics. 2010 Feb;9(2):327-35. doi: 10.1074/mcp.M900248-MCP200. Epub 2009 Nov 27. Mol Cell Proteomics. 2010. PMID: 19955079 Free PMC article.
-
Bayesian analysis of high-throughput quantitative measurement of protein-DNA interactions.PLoS One. 2011;6(11):e26105. doi: 10.1371/journal.pone.0026105. Epub 2011 Nov 1. PLoS One. 2011. PMID: 22069446 Free PMC article.
-
A DNA-Based Assay for Digoxin Detection.Biosensors (Basel). 2018 Mar 6;8(1):19. doi: 10.3390/bios8010019. Biosensors (Basel). 2018. PMID: 29509662 Free PMC article.
References
-
- Ptashne M, Gann A. Nature. 1997;386:569–577. - PubMed
-
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. Science. 2001;291:1304–1351. - PubMed
-
- Orlando V, Paro R. Cell. 1993;75:1187–1198. - PubMed
-
- Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, et al. Science. 2000;290:2306–2309. - PubMed
-
- Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, Brown PO. Nature. 2001;409:533–538. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

