RNA-binding proteins that inhibit RNA virus infection
- PMID: 17360619
- PMCID: PMC1805585
- DOI: 10.1073/pnas.0611617104
RNA-binding proteins that inhibit RNA virus infection
Abstract
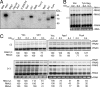
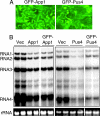
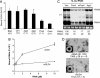
Arrays of >5,000 Saccharomyces cerevisiae proteins were screened to identify proteins that can preferentially bind a small RNA hairpin that contains a clamped adenine motif (CAM). A CAM is required for the replication of Brome Mosaic Virus (BMV), a plant-infecting RNA virus that can replicate in S. cerevisiae. Several hits were selected for further characterization in Nicotiana benthamiana. Pseudouridine Synthase 4 (Pus4) and the Actin Patch Protein 1 (App1) modestly reduced BMV genomic plus-strand RNA accumulation, but dramatically inhibited BMV systemic spread in plants. Pus4 also prevented the encapsidation of a BMV RNA in plants and the reassembly of BMV virions in vitro. These results demonstrate the feasibility of using proteome arrays to identify specific RNA-binding proteins for antiviral activities. Furthermore, the effects of Pus4 suggest that the CAM-containing RNA motif provides a regulatory link between RNA replication and encapsidation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Interactions between brome mosaic virus RNAs and cytoplasmic processing bodies.J Virol. 2007 Sep;81(18):9759-68. doi: 10.1128/JVI.00844-07. Epub 2007 Jul 3. J Virol. 2007. PMID: 17609284 Free PMC article.
-
The Lsm1-7-Pat1 complex promotes viral RNA translation and replication by differential mechanisms.RNA. 2015 Aug;21(8):1469-79. doi: 10.1261/rna.052209.115. Epub 2015 Jun 19. RNA. 2015. PMID: 26092942 Free PMC article.
-
Molecular studies on bromovirus capsid protein. VII. Selective packaging on BMV RNA4 by specific N-terminal arginine residuals.Virology. 2000 Sep 15;275(1):207-17. doi: 10.1006/viro.2000.0513. Virology. 2000. PMID: 11017800
-
The brome mosaic virus 3' untranslated sequence regulates RNA replication, recombination, and virion assembly.Virus Res. 2015 Aug 3;206:46-52. doi: 10.1016/j.virusres.2015.02.007. Epub 2015 Feb 14. Virus Res. 2015. PMID: 25687214 Review.
-
Bromovirus-induced remodeling of host membranes during viral RNA replication.Curr Opin Virol. 2014 Dec;9:104-10. doi: 10.1016/j.coviro.2014.09.018. Epub 2014 Oct 16. Curr Opin Virol. 2014. PMID: 25462441 Review.
Cited by
-
Overview of protein microarrays.Curr Protoc Protein Sci. 2013 Apr;Chapter 27(1):Unit 27.1. doi: 10.1002/0471140864.ps2701s72. Curr Protoc Protein Sci. 2013. PMID: 23546620 Free PMC article. Review.
-
Identification and characterization of the 480-kilodalton template-specific RNA-dependent RNA polymerase complex of red clover necrotic mosaic virus.J Virol. 2010 Jun;84(12):6070-81. doi: 10.1128/JVI.00054-10. Epub 2010 Apr 7. J Virol. 2010. PMID: 20375154 Free PMC article.
-
RNA Binding Proteins as Pioneer Determinants of Infection: Protective, Proviral, or Both?Viruses. 2021 Oct 28;13(11):2172. doi: 10.3390/v13112172. Viruses. 2021. PMID: 34834978 Free PMC article. Review.
-
A locking mechanism regulates RNA synthesis and host protein interaction by the hepatitis C virus polymerase.J Biol Chem. 2008 Jul 18;283(29):20535-46. doi: 10.1074/jbc.M801490200. Epub 2008 Apr 28. J Biol Chem. 2008. PMID: 18442978 Free PMC article.
-
Dual Roles of Host Zinc Finger Proteins in Viral RNA Regulation: Decay or Stabilization.Int J Mol Sci. 2024 Oct 17;25(20):11138. doi: 10.3390/ijms252011138. Int J Mol Sci. 2024. PMID: 39456919 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

