Extending the absorbing boundary method to fit dwell-time distributions of molecular motors with complex kinetic pathways
- PMID: 17360624
- PMCID: PMC1805548
- DOI: 10.1073/pnas.0611519104
Extending the absorbing boundary method to fit dwell-time distributions of molecular motors with complex kinetic pathways
Abstract
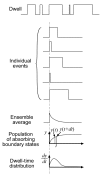
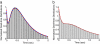
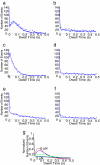
Dwell-time distributions, waiting-time distributions, and distributions of pause durations are widely reported for molecular motors based on single-molecule biophysical experiments. These distributions provide important information concerning the functional mechanisms of enzymes and their underlying kinetic and mechanical processes. We have extended the absorbing boundary method to simulate dwell-time distributions of complex kinetic schemes, which include cyclic, branching, and reverse transitions typically observed in molecular motors. This extended absorbing boundary method allows global fitting of dwell-time distributions for enzymes subject to different experimental conditions. We applied the extended absorbing boundary method to experimental dwell-time distributions of single-headed myosin V, and were able to use a single kinetic scheme to fit dwell-time distributions observed under different ligand concentrations and different directions of optical trap forces. The ability to use a single kinetic scheme to fit dwell-time distributions arising from a variety of experimental conditions is important for identifying a mechanochemical model of a molecular motor. This efficient method can be used to study dwell-time distributions for a broad class of molecular motors, including kinesin, RNA polymerase, helicase, F(1) ATPase, and to examine conformational dynamics of other enzymes such as ion channels.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Model for kinetics of myosin-V molecular motors.Biophys Chem. 2006 Apr 1;120(3):225-36. doi: 10.1016/j.bpc.2005.11.008. Epub 2005 Dec 28. Biophys Chem. 2006. PMID: 16386350
-
Maximum likelihood estimation of molecular motor kinetics from staircase dwell-time sequences.Biophys J. 2006 Aug 15;91(4):1156-68. doi: 10.1529/biophysj.105.079541. Epub 2006 May 5. Biophys J. 2006. PMID: 16679362 Free PMC article.
-
Dwell time distributions of the molecular motor myosin V.PLoS One. 2013;8(2):e55366. doi: 10.1371/journal.pone.0055366. Epub 2013 Feb 13. PLoS One. 2013. PMID: 23418440 Free PMC article.
-
Dwell time symmetry in random walks and molecular motors.Biophys J. 2007 Jun 1;92(11):3804-16. doi: 10.1529/biophysj.106.103044. Epub 2007 Mar 16. Biophys J. 2007. PMID: 17369422 Free PMC article. Review.
-
Molecular motors: a theorist's perspective.Annu Rev Phys Chem. 2007;58:675-95. doi: 10.1146/annurev.physchem.58.032806.104532. Annu Rev Phys Chem. 2007. PMID: 17163836 Review.
Cited by
-
DNA replication: In vitro single-molecule manipulation data analysis and models.Comput Struct Biotechnol J. 2021 Jun 24;19:3765-3778. doi: 10.1016/j.csbj.2021.06.032. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 34285777 Free PMC article. Review.
-
Mechanisms and topology determination of complex chemical and biological network systems from first-passage theoretical approach.J Chem Phys. 2013 Oct 14;139(14):144106. doi: 10.1063/1.4824392. J Chem Phys. 2013. PMID: 24116602 Free PMC article.
-
Power-law behavior of transcription factor dynamics at the single-molecule level implies a continuum affinity model.Nucleic Acids Res. 2021 Jul 9;49(12):6605-6620. doi: 10.1093/nar/gkab072. Nucleic Acids Res. 2021. PMID: 33592625 Free PMC article.
-
Quantitative analysis of DNA-looping kinetics from tethered particle motion experiments.Methods Enzymol. 2010;475:199-220. doi: 10.1016/S0076-6879(10)75009-6. Methods Enzymol. 2010. PMID: 20627159 Free PMC article.
-
Dynein-Inspired Multilane Exclusion Process with Open Boundary Conditions.Entropy (Basel). 2021 Oct 14;23(10):1343. doi: 10.3390/e23101343. Entropy (Basel). 2021. PMID: 34682067 Free PMC article.
References
-
- Liao J-C, Jeong Y-J, Kim D-E, Patel SS, Oster G. J Mol Biol. 2005;350:452–475. - PubMed
-
- Gardiner CW. Handbook of Stochastic Methods for Physics, Chemistry, and the Natural Sciences. Berlin: Springer; 1985.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

