Growth of novel protein structural data
- PMID: 17360626
- PMCID: PMC1802002
- DOI: 10.1073/pnas.0611678104
Growth of novel protein structural data
Abstract
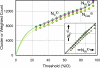
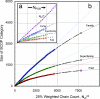
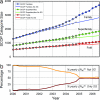
Contrary to popular assumption, the rate of growth of structural data has slowed, and the Protein Data Bank (PDB) has not been growing exponentially since 1995. Reaching such a dramatic conclusion requires careful measurement of growth of novel structures, which can be achieved by clustering entry sequences, or by using a novel index to down-weight entries with a higher number of sequence neighbors. These measures agree, and growth rates are very similar for entire PDB files, clusters, and weighted chains. The overall sizes of Structural Classification of Proteins (SCOP) categories (number of families, superfamilies, and folds) appear to be directly proportional to the number of deposited PDB files. Using our weighted chain count, which is most correlated to the change in the size of each SCOP category in any time period, shows that the rate of increase of SCOP categories is actually slowing down. This enables the final size of each of these SCOP categories to be predicted without examining or comparing protein structures. In the last 3 years, structures solved by structural genomics (SG) initiatives, especially the United States National Institutes of Health Protein Structure Initiative, have begun to redress the slowing growth of the PDB. Structures solved by SG are 3.8 times less sequence-redundant than typical PDB structures. Since mid-2004, SG programs have contributed half the novel structures measured by weighted chain counts. Our analysis does not rely on visual inspection of coordinate sets: it is done automatically, providing an accurate, up-to-date measure of the growth of novel protein structural data.
Conflict of interest statement
The author declares no conflict of interest.
Figures

References
-
- Kendrew JC, Dickerson RE, Strandberg BE, Hart RG, Davies DR, Phillips DC, Shore VC. Nature. 1961;185:422–427. - PubMed
-
- Kendrew JC. Sci Am. 1961;205:96–111. - PubMed
-
- Levitt M, Chothia C. Nature. 1976;261:552–558. - PubMed
-
- Bernstein FC, Koetzle TF, Williams GJ, Meyer EF, Jr, Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M. Eur J Biochem. 1977;80:319–324. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

