MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors
- PMID: 17360662
- PMCID: PMC1805579
- DOI: 10.1073/pnas.0611192104
MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors
Abstract
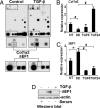
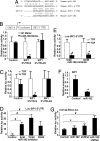
Key features of diabetic nephropathy (DN) include the accumulation of extracellular matrix proteins such as collagen 1-alpha 1 and -2 (Col1a1 and -2). Transforming growth factor beta1 (TGF-beta), a key regulator of these extracellular matrix genes, is increased in mesangial cells (MC) in DN. By microarray profiling, we noted that TGF-beta increased Col1a2 mRNA in mouse MC (MMC) but also decreased mRNA levels of an E-box repressor, deltaEF1. TGF-beta treatment or short hairpin RNAs targeting deltaEF1 increased enhancer activity of upstream E-box elements in the Col1a2 gene. TGF-beta also decreased the expression of Smad-interacting protein 1 (SIP1), another E-box repressor similar to deltaEF1. Interestingly, we noted that SIP1 is a target of microRNA-192 (miR-192), a key miR highly expressed in the kidney. miR-192 levels also were increased by TGF-beta in MMC. TGF-beta treatment or transfection with miR-192 decreased endogenous SIP1 expression as well as reporter activity of a SIP1 3' UTR-containing luciferase construct in MMC. Conversely, a miR-192 inhibitor enhanced the luciferase activity, confirming SIP1 to be a miR-192 target. Furthermore, miR-192 synergized with deltaEF1 short hairpin RNAs to increase Col1a2 E-box-luc activity. Importantly, the in vivo relevance was noted by the observation that miR-192 levels were enhanced significantly in glomeruli isolated from streptozotocin-injected diabetic mice as well as diabetic db/db mice relative to corresponding nondiabetic controls, in parallel with increased TGF-beta and Col1a2 levels. These results uncover a role for miRs in the kidney and DN in controlling TGF-beta-induced Col1a2 expression by down-regulating E-box repressors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Ziyadeh FN. Am J Kidney Dis. 1993;22:736–744. - PubMed
-
- Sharma K, Ziyadeh FN. Diabetes. 1995;44:1139–1146. - PubMed
-
- Kato M, Yuan H, Xu Z-G, Lanting L, Li S-L, Wang M, Hu MC-T, Reddy M, Natarajan R. J Am Soc Nephrol. 2006;17:3325–3335. - PubMed
-
- Roberts AB, McCune BK, Sporn MB. Kidney Int. 1992;41:557–559. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

