Single-site mutations in the carboxyltransferase domain of plastid acetyl-CoA carboxylase confer resistance to grass-specific herbicides
- PMID: 17360693
- PMCID: PMC1802000
- DOI: 10.1073/pnas.0611572104
Single-site mutations in the carboxyltransferase domain of plastid acetyl-CoA carboxylase confer resistance to grass-specific herbicides
Abstract
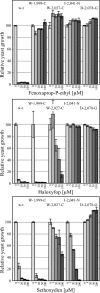
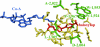
Grass weed populations resistant to aryloxyphenoxypropionate (APP) and cyclohexanedione herbicides that inhibit acetyl-CoA carboxylase (ACCase; EC 6.4.1.2) represent a major problem for sustainable agriculture. We investigated the molecular basis of resistance to ACCase-inhibiting herbicides for nine wild oat (Avena sterilis ssp. ludoviciana Durieu) populations from the northern grain-growing region of Australia. Five amino acid substitutions in plastid ACCase were correlated with herbicide resistance: Ile-1,781-Leu, Trp-1,999-Cys, Trp-2,027-Cys, Ile-2,041-Asn, and Asp-2,078-Gly (numbered according to the Alopecurus myosuroides plastid ACCase). An allele-specific PCR test was designed to determine the prevalence of these five mutations in wild oat populations suspected of harboring ACCase-related resistance with the result that, in most but not all cases, plant resistance was correlated with one (and only one) of the five mutations. We then showed, using a yeast gene-replacement system, that these single-site mutations also confer herbicide resistance to wheat plastid ACCase: Ile-1,781-Leu and Asp-2,078-Gly confer resistance to APPs and cyclohexanediones, Trp-2,027-Cys and Ile-2,041-Asn confer resistance to APPs, and Trp-1,999-Cys confers resistance only to fenoxaprop. These mutations are very likely to confer resistance to any grass weed species under selection imposed by the extensive agricultural use of the herbicides.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Delye C. Weed Sci. 2005;53:728–746.
-
- Brown AC, Moss SR, Wilson ZA, Field LM. Pest Biochem Physiol. 2002;72:160–168.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

