Tyrosine 3 of poliovirus terminal peptide VPg(3B) has an essential function in RNA replication in the context of its precursor protein, 3AB
- PMID: 17360746
- PMCID: PMC1900252
- DOI: 10.1128/JVI.02350-06
Tyrosine 3 of poliovirus terminal peptide VPg(3B) has an essential function in RNA replication in the context of its precursor protein, 3AB
Abstract
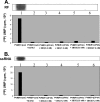
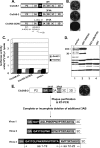
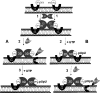
Poliovirus (PV) VPg is a genome-linked protein that is essential for the initiation of viral RNA replication. It has been well established that RNA replication is initiated when a molecule of UMP is covalently linked to the hydroxyl group of a tyrosine (Y3) in VPg by the viral RNA polymerase 3D(pol), but it is not yet known whether the substrate for uridylylation in vivo is the free peptide itself or one of its precursors. The aim of this study was to use complementation analyses to obtain information about the true in vivo substrate for uridylylation by 3D(pol). Previously, it was shown that a VPg mutant, in which tyrosine 3 and threonine 4 were replaced by phenylalanine and alanine (3F4A), respectively, was nonviable. We have now tested whether wild-type forms of proteins 3B, 3BC, 3BCD, 3AB, 3ABC, and P3 provided either in trans or in cis could rescue the replication defect of the VPg(3F4A) mutations in the PV polyprotein. Our results showed that proteins 3B, 3BC, 3BCD, and P3 were unable to complement the RNA replication defect in dicistronic PV or dicistronic luciferase replicons in vivo. However, cotranslation of the P3 precursor protein allowed rescue of RNA replication of the VPg(3F4A) mutant in an in vitro cell-free translation-RNA replication system, but only poor complementation was observed when 3BC, 3AB, 3BCD, or 3ABC proteins were cotranslated in the same assay. Interestingly, only protein 3AB but not 3B and 3BC, when provided in cis by insertion of a wild-type 3AB coding sequence between the P2 and P3 domains of the polyprotein, supported the replication of the mutated genome in vivo. Elimination of cleavage between 3A and 3B in the complementing 3AB protein, however, led to a complete lack of RNA replication. Our results suggest that (i) VPg has to be delivered to the replication complex in the form of a large protein precursor (P3) to be fully functional in replication; (ii) the replication complex formed during PV replication in vivo is essentially inaccessible to proteins provided in trans, even if the complementing protein is translated from a different cistron of the same RNA genome; (iii) 3AB is the most likely precursor of VPg; and (iv) Y3 of VPg has an essential function in RNA replication in the context of both VPg and 3AB.
Figures







References
-
- Agut, H., K. M. Kean, O. Fichot, J. Morasco, J. B. Flanegan, and M. Girard. 1989. A point mutation in the poliovirus polymerase gene determines a complementable temperature-sensitive defect of RNA replication. Virology 168:302-311. - PubMed
-
- Ambros, V., and D. Baltimore. 1978. Protein is linked to the 5′ end of poliovirus RNA by a phosphodiester linkage to tyrosine. J. Biol. Chem. 253:5263-5266. - PubMed
-
- Banerjee, R., and A. Dasgupta. 2001. Interaction of picornavirus 2C polypeptide with the viral negative-strand RNA. J. Gen. Virol. 82:2621-2627. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

