Simultaneous ex vivo quantification of antigen-specific CD4+ and CD8+ T cell responses using in vitro transcribed RNA
- PMID: 17361438
- PMCID: PMC11029841
- DOI: 10.1007/s00262-007-0302-7
Simultaneous ex vivo quantification of antigen-specific CD4+ and CD8+ T cell responses using in vitro transcribed RNA
Abstract
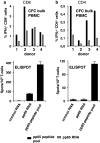
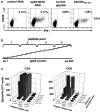
Assessment of antigen-specific T-cell responses has been greatly facilitated by development of ELISPOT and intracellular cytokine flow cytometry (CFC) assays. The use of autologous antigen presenting cells transfected with in vitro transcribed RNA as stimulators allows in principle quantification of antigen-specific T-cells independent of the knowledge of the epitopes. We describe here a cytokine secretion assay that enables simultaneous assessment of both antigen-specific CD4+ as well as CD8+ T-cells directly from clinical samples without the need for generation of dendritic cells. To this aim, bulk PBMCs were electroporated with RNA encoding the antigen fused to trafficking signal sequences derived from a MHC class I molecule and used as stimulators. With human cytomegalovirus (HCMV) phosphoprotein 65 (pp65) as antigen we show that for measuring ex vivo T-cell responses in ELISPOT and CFC such stimulators are superior or at least equivalent to a pool of overlapping peptides representing the entire pp65 sequence as well as to untagged pp65 encoding RNA. This approach avoids the time consuming generation of dendritic cells as immune stimulators and, in particular when used in the context of the CFC, is robust, broadly applicable and fast.
Figures


Similar articles
-
Identification of T cell epitopes by the use of rapidly generated mRNA fragments.J Immunol Methods. 2005 Apr;299(1-2):165-75. doi: 10.1016/j.jim.2005.02.004. Epub 2005 Mar 31. J Immunol Methods. 2005. PMID: 15914199
-
Use of a lentiviral vector encoding a HCMV-chimeric IE1-pp65 protein for epitope identification in HLA-Transgenic mice and for ex vivo stimulation and expansion of CD8(+) cytotoxic T cells from human peripheral blood cells.Hum Immunol. 2004 May;65(5):514-22. doi: 10.1016/j.humimm.2004.02.018. Hum Immunol. 2004. PMID: 15172452
-
Increased antigen presentation efficiency by coupling antigens to MHC class I trafficking signals.J Immunol. 2008 Jan 1;180(1):309-18. doi: 10.4049/jimmunol.180.1.309. J Immunol. 2008. PMID: 18097032
-
Progress made towards the development of a CMV peptide vaccine.Hum Immunol. 2004 May;65(5):544-9. doi: 10.1016/j.humimm.2004.02.005. Hum Immunol. 2004. PMID: 15172455 Review.
-
Dendritic cells, interleukin 12, and CD4+ lymphocytes in the initiation of class I-restricted reactivity to a tumor/self peptide.Crit Rev Immunol. 1998;18(1-2):87-98. doi: 10.1615/critrevimmunol.v18.i1-2.100. Crit Rev Immunol. 1998. PMID: 9419451 Review.
Cited by
-
A liposomal RNA vaccine inducing neoantigen-specific CD4+ T cells augments the antitumor activity of local radiotherapy in mice.Oncoimmunology. 2020 Jun 22;9(1):1771925. doi: 10.1080/2162402X.2020.1771925. Oncoimmunology. 2020. PMID: 32923128 Free PMC article.
-
A Trans-amplifying RNA Vaccine Strategy for Induction of Potent Protective Immunity.Mol Ther. 2020 Jan 8;28(1):119-128. doi: 10.1016/j.ymthe.2019.09.009. Epub 2019 Sep 12. Mol Ther. 2020. PMID: 31624015 Free PMC article.
-
Selective activation of trophoblast-specific PLAC1 in breast cancer by CCAAT/enhancer-binding protein beta (C/EBPbeta) isoform 2.J Biol Chem. 2009 Oct 16;284(42):28607-15. doi: 10.1074/jbc.M109.031120. Epub 2009 Aug 3. J Biol Chem. 2009. PMID: 19652226 Free PMC article.
-
Ionizable polymeric nanocarriers for the codelivery of bi-adjuvant and neoantigens in combination tumor immunotherapy.Bioact Mater. 2023 Aug;26:169-180. doi: 10.1016/j.bioactmat.2023.02.016. Epub 2023 Mar 3. Bioact Mater. 2023. PMID: 36883121 Free PMC article.
-
mRNA-based therapeutics--developing a new class of drugs.Nat Rev Drug Discov. 2014 Oct;13(10):759-80. doi: 10.1038/nrd4278. Epub 2014 Sep 19. Nat Rev Drug Discov. 2014. PMID: 25233993 Review.
References
-
- Clay TM, Hobeika AC, Mosca PJ, Lyerly HK, Morse MA. Assays for monitoring cellular immune responses to active immunotherapy of cancer. Clin Cancer Res. 2001;7:1127–1135. - PubMed
-
- Morse MA, Clay TM, Hobeika AC, Mosca PJ, Lyerly HK. Monitoring cellular immune responses to cancer immunotherapy. Curr Opin Mol Ther. 2001;3:45–52. - PubMed
-
- Brosterhus H, Brings S, Leyendeckers H, Manz RA, Miltenyi S, Radbruch A, Assenmacher M, Schmitz J. Enrichment and detection of live antigen-specific CD4(+) and CD8(+) T cells based on cytokine secretion. Eur J Immunol. 1999;29:4053–4059. doi: 10.1002/(SICI)1521-4141(199912)29:12<4053::AID-IMMU4053>3.0.CO;2-C. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

