The capsule polysaccharide structure and biogenesis for non-O1 Vibrio cholerae NRT36S: genes are embedded in the LPS region
- PMID: 17362509
- PMCID: PMC1847822
- DOI: 10.1186/1471-2180-7-20
The capsule polysaccharide structure and biogenesis for non-O1 Vibrio cholerae NRT36S: genes are embedded in the LPS region
Abstract
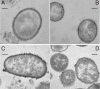
Background: In V. cholerae, the biogenesis of capsule polysaccharide is poorly understood. The elucidation of capsule structure and biogenesis is critical to understanding the evolution of surface polysaccharide and the internal relationship between the capsule and LPS in this species. V. cholerae serogroup O31 NRT36S, a human pathogen that produces a heat-stable enterotoxin (NAG-ST), is encapsulated. Here, we report the covalent structure and studies of the biogenesis of the capsule in V. cholerae NRT36S.
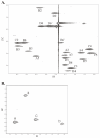
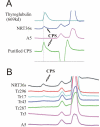
Results: The structure of the capsular (CPS) polysaccharide was determined by high resolution NMR spectroscopy and shown to be a complex structure with four residues in the repeating subunit. The gene cluster of capsule biogenesis was identified by transposon mutagenesis combined with whole genome sequencing data (GenBank accession DQ915177). The capsule gene cluster shared the same genetic locus as that of the O-antigen of lipopolysaccharide (LPS) biogenesis gene cluster. Other than V. cholerae O139, this is the first V. cholerae CPS for which a structure has been fully elucidated and the genetic locus responsible for biosynthesis identified.
Conclusion: The co-location of CPS and LPS biosynthesis genes was unexpected, and would provide a mechanism for simultaneous emergence of new O and K antigens in a single strain. This, in turn, may be a key element for V. cholerae to evolve new strains that can escape immunologic detection by host populations.
Figures
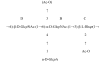



References
-
- Holmgren J, Svennerholm AM. Mechanisms of disease and immunity in cholera: a review. J Infect Dis. 1977;136 Suppl:S105–12. - PubMed
-
- Li M, Shimada T, Morris JG, Jr., Sulakvelidze A, Sozhamannan S. Evidence for the emergence of non-O1 and non-O139 Vibrio cholerae strains with pathogenic potential by exchange of O-antigen biosynthesis regions. Infect Immun. 2002;70:2441–2453. doi: 10.1128/IAI.70.5.2441-2453.2002. - DOI - PMC - PubMed
-
- Manning PA, Heuzenroeder MW, Yeadon J, Leavesley DI, Reeves PR, Rowley D. Molecular cloning and expression in Escherichia coli K-12 of the O antigens of the Inaba and Ogawa serotypes of the Vibrio cholerae O1 lipopolysaccharides and their potential for vaccine development. Infect Immun. 1986;53:272–277. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

