The human PINK1 locus is regulated in vivo by a non-coding natural antisense RNA during modulation of mitochondrial function
- PMID: 17362513
- PMCID: PMC1831481
- DOI: 10.1186/1471-2164-8-74
The human PINK1 locus is regulated in vivo by a non-coding natural antisense RNA during modulation of mitochondrial function
Abstract
Background: Mutations in the PTEN induced putative kinase 1 (PINK1) are implicated in early-onset Parkinson's disease. PINK1 is expressed abundantly in mitochondria rich tissues, such as skeletal muscle, where it plays a critical role determining mitochondrial structural integrity in Drosophila.
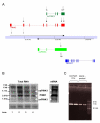
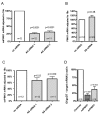
Results: Herein we characterize a novel splice variant of PINK1 (svPINK1) that is homologous to the C-terminus regulatory domain of the protein kinase. Naturally occurring non-coding antisense provides sophisticated mechanisms for diversifying genomes and we describe a human specific non-coding antisense expressed at the PINK1 locus (naPINK1). We further demonstrate that PINK1 varies in vivo when human skeletal muscle mitochondrial content is enhanced, supporting the idea that PINK1 has a physiological role in mitochondrion. The observation of concordant regulation of svPINK1 and naPINK1 during in vivo mitochondrial biogenesis was confirmed using RNAi, where selective targeting of naPINK1 results in loss of the PINK1 splice variant in neuronal cell lines.
Conclusion: Our data presents the first direct observation that a mammalian non-coding antisense molecule can positively influence the abundance of a cis-transcribed mRNA under physiological abundance conditions. While our analysis implies a possible human specific and dsRNA-mediated mechanism for stabilizing the expression of svPINK1, it also points to a broader genomic strategy for regulating a human disease locus and increases the complexity through which alterations in the regulation of the PINK1 locus could occur.
Figures

Similar articles
-
Existence of Pink1 antisense RNAs in mouse and their localization.Cytogenet Genome Res. 2009;126(3):259-70. doi: 10.1159/000251963. Epub 2010 Jan 6. Cytogenet Genome Res. 2009. PMID: 20068297
-
Altered regulation of the PINK1 locus: a link between type 2 diabetes and neurodegeneration?FASEB J. 2007 Nov;21(13):3653-65. doi: 10.1096/fj.07-8520com. Epub 2007 Jun 12. FASEB J. 2007. PMID: 17567565
-
Depletion of PINK1 affects mitochondrial metabolism, calcium homeostasis and energy maintenance.J Cell Sci. 2011 Apr 1;124(Pt 7):1115-25. doi: 10.1242/jcs.078303. Epub 2011 Mar 8. J Cell Sci. 2011. PMID: 21385841
-
Analysis of the regulatory and catalytic domains of PTEN-induced kinase-1 (PINK1).Hum Mutat. 2012 Oct;33(10):1408-22. doi: 10.1002/humu.22127. Epub 2012 Jul 5. Hum Mutat. 2012. PMID: 22644621 Review.
-
Mitochondrial dynamics, cell death and the pathogenesis of Parkinson's disease.Apoptosis. 2010 Nov;15(11):1336-53. doi: 10.1007/s10495-010-0465-0. Apoptosis. 2010. PMID: 20131004 Review.
Cited by
-
Epigenetic Modulation in Parkinson's Disease and Potential Treatment Therapies.Neurochem Res. 2021 Jul;46(7):1618-1626. doi: 10.1007/s11064-021-03334-w. Epub 2021 Apr 26. Neurochem Res. 2021. PMID: 33900517 Review.
-
The expression discrepancy and characteristics of long non-coding RNAs in peripheral blood leukocytes from amyotrophic lateral sclerosis patients.Mol Neurobiol. 2022 Jun;59(6):3678-3689. doi: 10.1007/s12035-022-02789-4. Epub 2022 Apr 2. Mol Neurobiol. 2022. PMID: 35364800
-
The Implications of the Long Non-Coding RNA NEAT1 in Non-Cancerous Diseases.Int J Mol Sci. 2019 Feb 1;20(3):627. doi: 10.3390/ijms20030627. Int J Mol Sci. 2019. PMID: 30717168 Free PMC article. Review.
-
Regulatory role of long non coding RNAs (lncRNAs) in neurological disorders: From novel biomarkers to promising therapeutic strategies.Asian J Pharm Sci. 2021 Sep;16(5):533-550. doi: 10.1016/j.ajps.2021.02.006. Epub 2021 Apr 2. Asian J Pharm Sci. 2021. PMID: 34849161 Free PMC article. Review.
-
Txnip balances metabolic and growth signaling via PTEN disulfide reduction.Proc Natl Acad Sci U S A. 2008 Mar 11;105(10):3921-6. doi: 10.1073/pnas.0800293105. Epub 2008 Mar 5. Proc Natl Acad Sci U S A. 2008. PMID: 18322014 Free PMC article.
References
-
- Valente EM, Abou-Sleiman PM, Caputo V, Muqit MM, Harvey K, Gispert S, Ali Z, Del Turco D, Bentivoglio AR, Healy DG, Albanese A, Nussbaum R, Gonzalez-Maldonado R, Deller T, Salvi S, Cortelli P, Gilks WP, Latchman DS, Harvey RJ, Dallapiccola B, Auburger G, Wood NW. Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science. 2004;304:1158–1160. doi: 10.1126/science.1096284. - DOI - PubMed
-
- Petit A, Kawarai T, Paitel E, Sanjo N, Maj M, Scheid M, Chen F, Gu Y, Hasegawa H, Salehi-Rad S, Wang L, Rogaeva E, Fraser P, Robinson B, St George-Hyslop P, Tandon A. Wild-type PINK1 prevents basal and induced neuronal apoptosis, a protective effect abrogated by Parkinson disease-related mutations. J Biol Chem. 2005;280:34025–34032. doi: 10.1074/jbc.M505143200. - DOI - PubMed
-
- Beilina A, Van Der Brug M, Ahmad R, Kesavapany S, Miller DW, Petsko GA, Cookson MR. Mutations in PTEN-induced putative kinase 1 associated with recessive parkinsonism have differential effects on protein stability. Proc Natl Acad Sci U S A. 2005;102:5703–5708. doi: 10.1073/pnas.0500617102. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

