Scavenger receptor-A functions in phagocytosis of E. coli by bone marrow dendritic cells
- PMID: 17362929
- PMCID: PMC1905149
- DOI: 10.1016/j.yexcr.2007.02.011
Scavenger receptor-A functions in phagocytosis of E. coli by bone marrow dendritic cells
Abstract
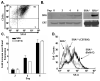
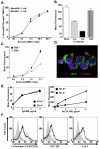
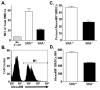
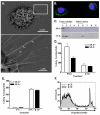
Class-A scavenger receptors (SR-A) are cellular pattern recognition receptors that bind and traffic a variety of endogenous and microbial ligands. However, despite an emerging role for SR-A as a contributor to the innate immune system, little is known of the regulation or function of SR-A on dendritic cells (DCs). Here we show that SR-A expression is upregulated during murine DC differentiation and that SR-A expression levels correlate with the expression of the murine DC marker CD11c. Using bone marrow-derived DCs (BMDCs) from SR-A knockout (SR-A(-/-)) mice, we investigated the contribution of SR-A to BMDC particulate phagocytosis. Functional analyses demonstrated that SR-A is a critical phagocytic receptor for BMDC internalization of the gram-negative bacteria E. coli. SR-A(-/-) BMDCs were impaired in their ability to phagocytose bacteria, and this deficit varied with the bacteria:BMDC cell ratio. Microscopic and biochemical analyses revealed that SR-A is broadly distributed on the surface of BMDCs and is not physically associated with lipid rafts. However, cholesterol depletion demonstrated dependence of SR-A-mediated phagocytosis upon lipid rafts. These data demonstrate a functional contribution for SR-A in the BMDC phagocytic pathway.
Figures

Similar articles
-
Macrophage class A scavenger receptor-mediated phagocytosis of Escherichia coli: role of cell heterogeneity, microbial strain, and culture conditions in vitro.Infect Immun. 2000 Apr;68(4):1953-63. doi: 10.1128/IAI.68.4.1953-1963.2000. Infect Immun. 2000. PMID: 10722588 Free PMC article.
-
Impact of culture medium on maturation of bone marrow-derived murine dendritic cells via the aryl hydrocarbon receptor.Mol Immunol. 2012 May;51(1):42-50. doi: 10.1016/j.molimm.2012.02.005. Epub 2012 Feb 28. Mol Immunol. 2012. PMID: 22377453
-
Modulation of dendritic cell differentiation in the bone marrow mediates sustained immunosuppression after polymicrobial sepsis.J Immunol. 2011 Jan 15;186(2):977-86. doi: 10.4049/jimmunol.1001147. Epub 2010 Dec 15. J Immunol. 2011. PMID: 21160046
-
[The role of the class A scavenger receptors, SR-A and MARCO, in the immune system. Part 2. Contribution to recognition and phagocytosis of pathogens as well as induction of immune response].Postepy Hig Med Dosw (Online). 2012 Feb 29;66:120-31. doi: 10.5604/17322693.984080. Postepy Hig Med Dosw (Online). 2012. PMID: 22470186 Review. Polish.
-
Bacterial antigen delivery systems: phagocytic processing of bacterial antigens for MHC-I and MHC-II presentation to T cells.Behring Inst Mitt. 1997 Feb;(98):197-211. Behring Inst Mitt. 1997. PMID: 9382741 Review.
Cited by
-
Phagocytosis of live versus killed or fluorescently labeled bacteria by macrophages differ in both magnitude and receptor specificity.Immunol Cell Biol. 2017 May;95(5):424-435. doi: 10.1038/icb.2016.112. Epub 2016 Nov 9. Immunol Cell Biol. 2017. PMID: 27826145
-
Pivotal Advance: Toll-like receptor regulation of scavenger receptor-A-mediated phagocytosis.J Leukoc Biol. 2009 Apr;85(4):595-605. doi: 10.1189/jlb.1008631. Epub 2008 Dec 26. J Leukoc Biol. 2009. PMID: 19112093 Free PMC article.
-
Cisplatin increases immune activity of monocytes and cytotoxic T-cells in a murine model of epithelial ovarian cancer.Transl Oncol. 2021 Dec;14(12):101217. doi: 10.1016/j.tranon.2021.101217. Epub 2021 Sep 14. Transl Oncol. 2021. PMID: 34530192 Free PMC article.
-
A co-assembly platform engaging macrophage scavenger receptor A for lysosome-targeting protein degradation.Nat Commun. 2024 Feb 23;15(1):1663. doi: 10.1038/s41467-024-46130-0. Nat Commun. 2024. PMID: 38396109 Free PMC article.
-
Implication of scavenger receptors in the interactions between diesel exhaust particles and immature or mature dendritic cells.Part Fibre Toxicol. 2009 Mar 13;6:9. doi: 10.1186/1743-8977-6-9. Part Fibre Toxicol. 2009. PMID: 19284653 Free PMC article.
References
-
- Brown MS, Goldstein JL. Lipoprotein metabolism in the macrophage: implications for cholesterol deposition in atherosclerosis. Annu Rev Biochem. 1983;52:223–61. - PubMed
-
- Krieger M, Herz J. Structures and functions of multiligand lipoprotein receptors: macrophage scavenger receptors and LDL receptor-related protein (LRP) Annu Rev Biochem. 1994;63:601–37. - PubMed
-
- Fraser I, Hughes D, Gordon S. Divalent cation-independent macrophage adhesion inhibited by monoclonal antibody to murine scavenger receptor. Nature. 1993;364:343–6. - PubMed
-
- Platt N, Haworth R, Darley L, Gordon S. The many roles of the class A macrophage scavenger receptor. Int Rev Cytol. 2002;212:1–40. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

