Notch signals positively regulate activity of the mTOR pathway in T-cell acute lymphoblastic leukemia
- PMID: 17363738
- PMCID: PMC1896117
- DOI: 10.1182/blood-2006-08-039883
Notch signals positively regulate activity of the mTOR pathway in T-cell acute lymphoblastic leukemia
Abstract
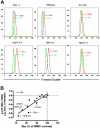
Constitutive Notch activation is required for the proliferation of a subgroup of T-cell acute lymphoblastic leukemia (T-ALL). Downstream pathways that transmit pro-oncogenic signals are not well characterized. To identify these pathways, protein microarrays were used to profile the phosphorylation state of 108 epitopes on 82 distinct signaling proteins in a panel of 13 T-cell leukemia cell lines treated with a gamma-secretase inhibitor (GSI) to inhibit Notch signals. The microarray screen detected GSI-induced hypophosphorylation of multiple signaling proteins in the mTOR pathway. This effect was rescued by expression of the intracellular domain of Notch and mimicked by dominant negative MAML1, confirming Notch specificity. Withdrawal of Notch signals prevented stimulation of the mTOR pathway by mitogenic factors. These findings collectively suggest that the mTOR pathway is positively regulated by Notch in T-ALL cells. The effect of GSI on the mTOR pathway was independent of changes in phosphatidylinositol-3 kinase and Akt activity, but was rescued by expression of c-Myc, a direct transcriptional target of Notch, implicating c-Myc as an intermediary between Notch and mTOR. T-ALL cell growth was suppressed in a highly synergistic manner by simultaneous treatment with the mTOR inhibitor rapamycin and GSI, which represents a rational drug combination for treating this aggressive human malignancy.
Figures





References
-
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. - PubMed
-
- Weng AP, Aster JC. Multiple niches for Notch in cancer: context is everything. Curr Opin Genet Dev. 2004;14:48–54. - PubMed
-
- Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. - PubMed
-
- Leong KG, Karsan A. Recent insights into the role of Notch signaling in tumorigenesis. Blood. 2006;107:2223–2233. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

