Mus81 cleavage of Holliday junctions: a failsafe for processing meiotic recombination intermediates?
- PMID: 17363897
- PMCID: PMC1847671
- DOI: 10.1038/sj.emboj.7601645
Mus81 cleavage of Holliday junctions: a failsafe for processing meiotic recombination intermediates?
Abstract
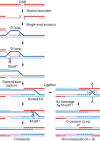
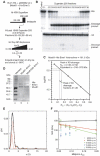
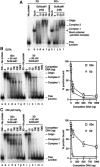
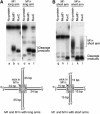
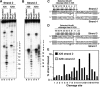
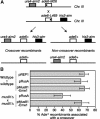
The Holliday junction (HJ) is a central intermediate of homologous recombination. Its cleavage is critical for the formation of crossover recombinants during meiosis, which in turn helps to establish chiasmata and promote genetic diversity. Enzymes that cleave HJs, called HJ resolvases, have been identified in all domains of life except eukaryotic nuclei. Controversially, the Mus81-Eme1 endonuclease has been proposed to be an example of a eukaryotic nuclear resolvase. However, hitherto little or no HJ cleavage has been detected in recombinant preparations of Mus81-Eme1. Here, we report the purification of active forms of recombinant Schizosaccharomyces pombe Mus81-Eme1 and Saccharomyces cerevisiae Mus81-Mms4, which display robust HJ cleavage in vitro, which, in the case of Mus81-Eme1, is as good as the archetypal HJ resolvase RuvC in single turnover kinetic analysis. We also present genetic evidence that suggests that this activity might be utilised as a back-up to Mus81-Eme1's main activity of cleaving nicked HJs during meiosis in S. pombe.
Figures

Similar articles
-
Efficient second strand cleavage during Holliday junction resolution by RuvC requires both increased junction flexibility and an exposed 5' phosphate.PLoS One. 2009;4(4):e5347. doi: 10.1371/journal.pone.0005347. Epub 2009 Apr 28. PLoS One. 2009. PMID: 19399178 Free PMC article.
-
Generating crossovers by resolution of nicked Holliday junctions: a role for Mus81-Eme1 in meiosis.Mol Cell. 2003 Sep;12(3):761-74. doi: 10.1016/s1097-2765(03)00343-5. Mol Cell. 2003. PMID: 14527420
-
Mus81-Mms4 functions as a single heterodimer to cleave nicked intermediates in recombinational DNA repair.Mol Cell Biol. 2012 Aug;32(15):3065-80. doi: 10.1128/MCB.00547-12. Epub 2012 May 29. Mol Cell Biol. 2012. PMID: 22645308 Free PMC article.
-
Holliday junction resolvases.Cold Spring Harb Perspect Biol. 2014 Sep 2;6(9):a023192. doi: 10.1101/cshperspect.a023192. Cold Spring Harb Perspect Biol. 2014. PMID: 25183833 Free PMC article. Review.
-
Recombination: Holliday junction resolution and crossover formation.Curr Biol. 2004 Jan 20;14(2):R56-8. doi: 10.1016/j.cub.2003.12.043. Curr Biol. 2004. PMID: 14738748 Review.
Cited by
-
Mms22 preserves genomic integrity during DNA replication in Schizosaccharomyces pombe.Genetics. 2007 Sep;177(1):47-61. doi: 10.1534/genetics.107.077255. Epub 2007 Jul 29. Genetics. 2007. PMID: 17660542 Free PMC article.
-
Expression of MUS81 Mediates the Sensitivity of Castration-Resistant Prostate Cancer to Olaparib.J Immunol Res. 2022 Jul 21;2022:4065580. doi: 10.1155/2022/4065580. eCollection 2022. J Immunol Res. 2022. PMID: 35910852 Free PMC article.
-
Cleavage mechanism of human Mus81-Eme1 acting on Holliday-junction structures.Proc Natl Acad Sci U S A. 2008 Mar 11;105(10):3757-62. doi: 10.1073/pnas.0710291105. Epub 2008 Feb 29. Proc Natl Acad Sci U S A. 2008. PMID: 18310322 Free PMC article.
-
Sgs1 RecQ helicase inhibits survival of Saccharomyces cerevisiae cells lacking telomerase and homologous recombination.J Biol Chem. 2008 Oct 31;283(44):29847-58. doi: 10.1074/jbc.M804760200. Epub 2008 Aug 29. J Biol Chem. 2008. PMID: 18757364 Free PMC article.
-
Increased meiotic crossovers and reduced genome stability in absence of Schizosaccharomyces pombe Rad16 (XPF).Genetics. 2014 Dec;198(4):1457-72. doi: 10.1534/genetics.114.171355. Epub 2014 Oct 6. Genetics. 2014. PMID: 25293972 Free PMC article.
References
-
- Abraham J, Lemmers B, Hande MP, Moynahan ME, Chahwan C, Ciccia A, Essers J, Hanada K, Chahwan R, Khaw AK, McPherson P, Shehabeldin A, Laister R, Arrowsmith C, Kanaar R, West SC, Jasin M, Hakem R (2003) Eme1 is involved in DNA damage processing and maintenance of genomic stability in mammalian cells. EMBO J 22: 6137–6147 - PMC - PubMed
-
- Allers T, Lichten M (2001) Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106: 47–57 - PubMed
-
- Boddy MN, Gaillard P-HL, McDonald WH, Shanahan P, Yates JR III, Russell P (2001) Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell 107: 537–548 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

