The human Nup107-160 nuclear pore subcomplex contributes to proper kinetochore functions
- PMID: 17363900
- PMCID: PMC1847668
- DOI: 10.1038/sj.emboj.7601642
The human Nup107-160 nuclear pore subcomplex contributes to proper kinetochore functions
Abstract
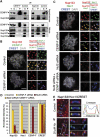
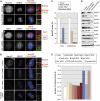
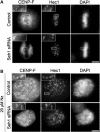
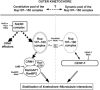
We previously demonstrated that a fraction of the human Nup107-160 nuclear pore subcomplex is recruited to kinetochores at the onset of mitosis. However, the molecular determinants for its kinetochore targeting and the functional significance of this localization were not investigated. Here, we show that the Nup107-160 complex interacts with CENP-F, but that CENP-F only moderately contributes to its targeting to kinetochores. In addition, we show that the recruitment of the Nup107-160 complex to kinetochores mainly depends on the Ndc80 complex. We further demonstrate that efficient depletion of the Nup107-160 complex from kinetochores, achieved either by combining siRNAs targeting several of its subunits excluding Seh1, or by depleting Seh1 alone, induces a mitotic delay. Further analysis of Seh1-depleted cells revealed impaired chromosome congression, reduced kinetochore tension and kinetochore-microtubule attachment defects. Finally, we show that the presence of the Nup107-160 complex at kinetochores is required for the recruitment of Crm1 and RanGAP1-RanBP2 to these structures. Together, our data thus provide the first molecular clues underlying the function of the human Nup107-160 complex at kinetochores.
Figures




References
-
- Arnaoutov A, Azuma Y, Ribbeck K, Joseph J, Boyarchuk Y, Karpova T, McNally J, Dasso M (2005) Crm1 is a mitotic effector of Ran-GTP in somatic cells. Nat Cell Biol 7: 626–632 - PubMed
-
- Arnaoutov A, Dasso M (2003) The Ran GTPase regulates kinetochore function. Dev Cell 5: 99–111 - PubMed
-
- Arnaoutov A, Dasso M (2005) Ran-GTP regulates kinetochore attachment in somatic cells. Cell Cycle 4: 1161–1165 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

