An alternative branch of the nonsense-mediated decay pathway
- PMID: 17363904
- PMCID: PMC1847659
- DOI: 10.1038/sj.emboj.7601628
An alternative branch of the nonsense-mediated decay pathway
Abstract
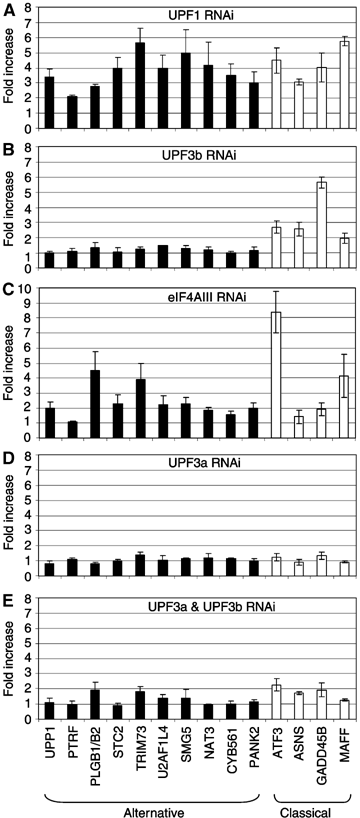
The T-cell receptor (TCR) locus undergoes programmed rearrangements that frequently generate premature termination codons (PTCs). The PTC-bearing transcripts derived from such nonproductively rearranged genes are dramatically downregulated by the nonsense-mediated decay (NMD) pathway. Here, we show that depletion of the NMD factor UPF3b does not impair TCRbeta NMD, thereby distinguishing it from classical NMD. Depletion of the related factor UPF3a, by itself or in combination with UPF3b, also has no effect on TCRbeta NMD. Mapping experiments revealed the identity of TCRbeta sequences that elicit a switch to UPF3b dependence. This regulation is not a peculiarity of TCRbeta, as we identified many wild-type genes, including one essential for NMD, that transcribe NMD-targeted mRNAs whose downregulation is little or not affected by UPF3a and UPF3b depletion. We propose that we have uncovered an alternative branch of the NMD pathway that not only degrades aberrant mRNAs but also regulates normal mRNAs, including one that participates in a negative feedback loop controlling the magnitude of NMD.
Figures




References
-
- Bruce SR, Wilkinson MF (2003) Nonsense mediated decay: a surveillance pathway that detects faulty TCR and BCR. In Recent Res Devel in Immunity, Pandalai SG (ed) Vol I, pp 1–12. Kerala, India: Research Signpost
-
- Buhler M, Steiner S, Mohn F, Paillusson A, Muhlemann O (2006) EJC-independent degradation of nonsense immunoglobulin-mu mRNA depends on 3′ UTR length. Nat Struct Mol Biol 13: 462–464 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

