Orientation Determination of Membrane-Disruptive Proteins Using Powder Samples and Rotational Diffusion: A Simple Solid-State NMR Approach
- PMID: 17364006
- PMCID: PMC1826912
- DOI: 10.1016/j.cplett.2006.10.067
Orientation Determination of Membrane-Disruptive Proteins Using Powder Samples and Rotational Diffusion: A Simple Solid-State NMR Approach
Abstract
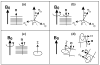
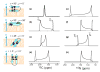
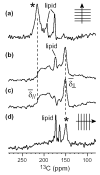
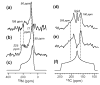
The orientation of membrane proteins undergoing fast uniaxial rotation around the bilayer normal can be determined without macroscopic alignment. We show that the motionally averaged powder spectra exhibit their 0° frequency, [Formula: see text], at the same position as the peak of an aligned sample with the alignment axis parallel to the magnetic field. This equivalence is exploited to determine the orientation of a β-sheet antimicrobial peptide not amenable to macroscopic alignment, using (13)CO and (15)N chemical shifts from powder spectra. This powder sample approach permits orientation determination of naturally membrane-disruptive proteins in diverse environments and under magic-angle spinning.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
