A mathematical model for neutrophil gradient sensing and polarization
- PMID: 17367201
- PMCID: PMC1828701
- DOI: 10.1371/journal.pcbi.0030036
A mathematical model for neutrophil gradient sensing and polarization
Abstract
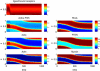
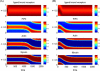
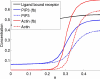
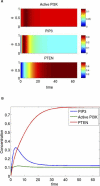
Directed cell migration in response to chemical cues, also known as chemotaxis, is an important physiological process involved in wound healing, foraging, and the immune response. Cell migration requires the simultaneous formation of actin polymers at the leading edge and actomyosin complexes at the sides and back of the cell. An unresolved question in eukaryotic chemotaxis is how the same chemoattractant signal determines both the cell's front and back. Recent experimental studies have begun to reveal the biochemical mechanisms necessary for this polarized cellular response. We propose a mathematical model of neutrophil gradient sensing and polarization based on experimentally characterized biochemical mechanisms. The model demonstrates that the known dynamics for Rho GTPase and phosphatidylinositol-3-kinase (PI3K) activation are sufficient for both gradient sensing and polarization. In particular, the model demonstrates that these mechanisms can correctly localize the "front" and "rear" pathways in response to both uniform concentrations and gradients of chemical attractants, including in actin-inhibited cells. Furthermore, the model predictions are robust to the values of many parameters. A key result of the model is the proposed coincidence circuit involving PI3K and Ras that obviates the need for the "global inhibitors" proposed, though never experimentally verified, in many previous mathematical models of eukaryotic chemotaxis. Finally, experiments are proposed to (in)validate this model and further our understanding of neutrophil chemotaxis.
Conflict of interest statement
Figures









References
-
- Manahan C, Iglesias P, Long Y, Deverotes P. Chemoattractant signaling in dictyostelium discoideum. Annu Rev Cell Dev Biol. 2004;20:223–253. - PubMed
-
- van Haastert P, Devreotes P. Chemotaxis: Signalling the way forward. Nat Rev Mol Cell Biol. 2004;5:626–634. - PubMed
-
- Li S, Guan J, Chein S. Biochemistry and biomechanics of cell motility. Annu Rev Biomed Eng. 2005;7:105–150. - PubMed
-
- Schiffmann E. Leukocyte chemotaxis. Annu Rev Physiol. 1982;44:553–568. - PubMed
-
- Deverotes P, Zigmond S. Chemotaxis in eukaryotic cells: A focus on leukocytes and Dictyostelium . Annu Rev Cell Biol. 1988;4:649–686. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

