Reactome: a knowledge base of biologic pathways and processes
- PMID: 17367534
- PMCID: PMC1868929
- DOI: 10.1186/gb-2007-8-3-r39
Reactome: a knowledge base of biologic pathways and processes
Erratum in
- Genome Biol. 2009 Feb 4;10(2):402. Joshi-Tope, Geeta [removed]
Abstract
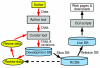
Reactome http://www.reactome.org, an online curated resource for human pathway data, provides infrastructure for computation across the biologic reaction network. We use Reactome to infer equivalent reactions in multiple nonhuman species, and present data on the reliability of these inferred reactions for the distantly related eukaryote Saccharomyces cerevisiae. Finally, we describe the use of Reactome both as a learning resource and as a computational tool to aid in the interpretation of microarrays and similar large-scale datasets.
Figures




References
-
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E. et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

