The ring between ring fingers (RBR) protein family
- PMID: 17367545
- PMCID: PMC1868946
- DOI: 10.1186/gb-2007-8-3-209
The ring between ring fingers (RBR) protein family
Abstract
Proteins of the ring between ring fingers (RBR)-domain family are characterized by three groups of specifically clustered (typically eight) cysteine and histidine residues. Whereas the amino-terminal ring domain (N-RING) binds two zinc ions and folds into a classical cross-brace ring finger, the carboxy-terminal ring domain (C-RING) involves only one zinc ion. The three-dimensional structure of the central ring domain, the IBR domain, is still unsolved. About 400 genes coding for RBR proteins have been identified in the genomes of uni- and multicellular eukaryotes and some of their viruses, but the family has not been found in archaea or bacteria. The RBR proteins are classified into 15 major subfamilies (besides some orphan cases) by the phylogenetic relationships of the RBR segments and the conservation of their sequence architecture. The RBR domain mediates protein-protein interactions and a subset of RBR proteins has been shown to function as E3 ubiquitin ligases. RBR proteins have attracted interest because of their involvement in diseases such as parkinsonism, dementia with Lewy bodies, and Alzheimer's disease, and in susceptibility to some intracellular bacterial pathogens. Here, we present an overview of the RBR-domain containing proteins and their subcellular localization, additional domains, function, specificity, and regulation.
Figures

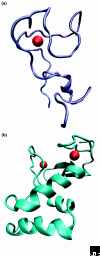

References
-
- Marin I, Ferrus A. Comparative genomics of the RBR family, including the Parkinson's disease-related gene parkin and the genes of the Ariadne subfamily. Mol Biol Evol. 2002;19:2039–2050. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

