Identification and characterization of genes encoding a putative ABC-type transporter essential for utilization of gamma-hexachlorocyclohexane in Sphingobium japonicum UT26
- PMID: 17369300
- PMCID: PMC1913331
- DOI: 10.1128/JB.01883-06
Identification and characterization of genes encoding a putative ABC-type transporter essential for utilization of gamma-hexachlorocyclohexane in Sphingobium japonicum UT26
Abstract
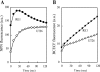
Sphingobium japonicum UT26 utilizes gamma-hexachlorocyclohexane (gamma-HCH) as its sole source of carbon and energy. In our previous studies, we cloned and characterized genes encoding enzymes for the conversion of gamma-HCH to beta-ketoadipate in UT26. In this study, we analyzed a mutant obtained by transposon mutagenesis and identified and characterized new genes encoding a putative ABC-type transporter essential for the utilization of gamma-HCH in strain UT26. This putative ABC transporter consists of four components, permease, ATPase, periplasmic protein, and lipoprotein, encoded by linK, linL, linM, and linN, respectively. Mutation and complementation analyses indicated that all the linKLMN genes are required, probably as a set, for gamma-HCH utilization in UT26. Furthermore, the mutant cells deficient in this putative ABC transporter showed (i) higher gamma-HCH degradation activity and greater accumulation of the toxic dead-end product 2,5-dichlorophenol (2,5-DCP), (ii) higher sensitivity to 2,5-DCP itself, and (iii) higher permeability of hydrophobic compounds than the wild-type cells. These results strongly suggested that LinKLMN are involved in gamma-HCH utilization by controlling membrane hydrophobicity. This study clearly demonstrated that a cellular factor besides catabolic enzymes and transcriptional regulators is essential for utilization of xenobiotic compounds in bacterial cells.
Figures




References
-
- Biemans-Oldehinkel, E., M. K. Doeven, and B. Poolman. 2006. ABC transporter architecture and regulatory roles of accessory domains. FEBS Lett. 580:1023-1035. - PubMed
-
- Camara, B., C. Herrera, M. Gonzalez, E. Couve, B. Hofer, and M. Seeger. 2004. From PCBs to highly toxic metabolites by the biphenyl pathway. Environ. Microbiol. 6:842-850. - PubMed
-
- Cassidy, M. B., H. Lee, J. T. Trevors, and R. B. Zablotowicz. 1999. Chlorophenol and nitrophenol metabolism by Sphingomonas sp. UG30. J. Ind. Microbiol. Biotechnol. 23:232-241. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

