Global analysis of exon creation versus loss and the role of alternative splicing in 17 vertebrate genomes
- PMID: 17369312
- PMCID: PMC1852814
- DOI: 10.1261/rna.325107
Global analysis of exon creation versus loss and the role of alternative splicing in 17 vertebrate genomes
Abstract
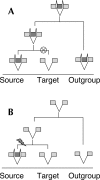
Association of alternative splicing (AS) with accelerated rates of exon evolution in some organisms has recently aroused widespread interest in its role in evolution of eukaryotic gene structure. Previous studies were limited to analysis of exon creation or lost events in mouse and/or human only. Our multigenome approach provides a way for (1) distinguishing creation and loss events on the large scale; (2) uncovering details of the evolutionary mechanisms involved; (3) estimating the corresponding rates over a wide range of evolutionary times and organisms; and (4) assessing the impact of AS on those evolutionary rates. We use previously unpublished independent analyses of alternative splicing in five species (human, mouse, dog, cow, and zebrafish) from the ASAP database combined with genomewide multiple alignment of 17 genomes to analyze exon creation and loss of both constitutively and alternatively spliced exons in mammals, fish, and birds. Our analysis provides a comprehensive database of exon creation and loss events over 360 million years of vertebrate evolution, including tens of thousands of alternative and constitutive exons. We find that exon inclusion level is inversely related to the rate of exon creation. In addition, we provide a detailed in-depth analysis of mechanisms of exon creation and loss, which suggests that a large fraction of nonrepetitive created exons are results of ab initio creation from purely intronic sequences. Our data indicate an important role for alternative splicing in creation of new exons and provide a useful novel database resource for future genome evolution research.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
