Maternal microRNAs are essential for mouse zygotic development
- PMID: 17369397
- PMCID: PMC1820938
- DOI: 10.1101/gad.418707
Maternal microRNAs are essential for mouse zygotic development
Abstract
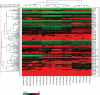
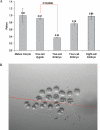
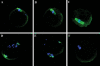
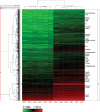
MicroRNAs (miRNAs) have important roles in diverse cellular processes, but little is known about their identity and functions during early mammalian development. Here, we show the effects of the loss of maternal inheritance of miRNAs following specific deletion of Dicer from growing oocytes. The mutant mature oocytes were almost entirely depleted of all miRNAs, and they failed to progress through the first cell division, probably because of disorganized spindle formation. By comparing single-cell cDNA microarray profiles of control and mutant oocytes, our data are compatible with the notion that a large proportion of the maternal genes are directly or indirectly under the control of miRNAs, which demonstrates that the maternal miRNAs are essential for the earliest stages of mouse embryonic development.
Figures
References
-
- Amanai M., Brahmajosyula M., Perry A.C., Brahmajosyula M., Perry A.C., Perry A.C. A restricted role for sperm-borne microRNAs in mammalian fertilization. Biol. Reprod. 2006;75:877–884. - PubMed
-
- Bagga S., Bracht J., Hunter S., Massirer K., Holtz J., Eachus R., Pasquinelli A.E., Bracht J., Hunter S., Massirer K., Holtz J., Eachus R., Pasquinelli A.E., Hunter S., Massirer K., Holtz J., Eachus R., Pasquinelli A.E., Massirer K., Holtz J., Eachus R., Pasquinelli A.E., Holtz J., Eachus R., Pasquinelli A.E., Eachus R., Pasquinelli A.E., Pasquinelli A.E. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. - PubMed
-
- Bartel D.P. MicroRNAs: Fenomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
-
- Bernstein E., Kim S.Y., Carmell M.A., Murchison E.P., Alcorn H., Li M.Z., Mills A.A., Elledge S.J., Anderson K.V., Hannon G.J., Kim S.Y., Carmell M.A., Murchison E.P., Alcorn H., Li M.Z., Mills A.A., Elledge S.J., Anderson K.V., Hannon G.J., Carmell M.A., Murchison E.P., Alcorn H., Li M.Z., Mills A.A., Elledge S.J., Anderson K.V., Hannon G.J., Murchison E.P., Alcorn H., Li M.Z., Mills A.A., Elledge S.J., Anderson K.V., Hannon G.J., Alcorn H., Li M.Z., Mills A.A., Elledge S.J., Anderson K.V., Hannon G.J., Li M.Z., Mills A.A., Elledge S.J., Anderson K.V., Hannon G.J., Mills A.A., Elledge S.J., Anderson K.V., Hannon G.J., Elledge S.J., Anderson K.V., Hannon G.J., Anderson K.V., Hannon G.J., Hannon G.J. Dicer is essential for mouse development. Nat. Genet. 2003;35:215–217. - PubMed
-
- Chen C., Ridzon D.A., Broomer A.J., Zhou Z., Lee D.H., Nguyen J.T., Barbisin M., Xu N.L., Mahuvakar V.R., Andersen M.R., Ridzon D.A., Broomer A.J., Zhou Z., Lee D.H., Nguyen J.T., Barbisin M., Xu N.L., Mahuvakar V.R., Andersen M.R., Broomer A.J., Zhou Z., Lee D.H., Nguyen J.T., Barbisin M., Xu N.L., Mahuvakar V.R., Andersen M.R., Zhou Z., Lee D.H., Nguyen J.T., Barbisin M., Xu N.L., Mahuvakar V.R., Andersen M.R., Lee D.H., Nguyen J.T., Barbisin M., Xu N.L., Mahuvakar V.R., Andersen M.R., Nguyen J.T., Barbisin M., Xu N.L., Mahuvakar V.R., Andersen M.R., Barbisin M., Xu N.L., Mahuvakar V.R., Andersen M.R., Xu N.L., Mahuvakar V.R., Andersen M.R., Mahuvakar V.R., Andersen M.R., Andersen M.R., et al. Real-time quantification of microRNAs by stem-loop RT–PCR. Nucleic Acids Res. 2005;33:e179. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
