Replication fork stalling and cell cycle arrest in UV-irradiated Escherichia coli
- PMID: 17369400
- PMCID: PMC1820941
- DOI: 10.1101/gad.417607
Replication fork stalling and cell cycle arrest in UV-irradiated Escherichia coli
Abstract
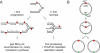
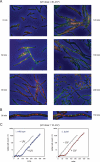
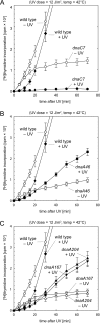
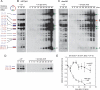
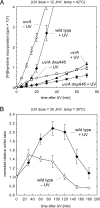
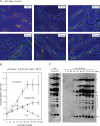
Faithful duplication of the genome relies on the ability to cope with an imperfect template. We investigated replication of UV-damaged DNA in Escherichia coli and found that ongoing replication stops for at least 15-20 min before resuming. Undamaged origins of replication (oriC) continue to fire at the normal rate and in a DnaA-dependent manner. UV irradiation also induces substantial DnaA-independent replication. These two factors add substantially to the DNA synthesis detected after irradiation and together mask the delay in the progression of pre-existing forks in assays measuring net synthesis. All DNA synthesis after UV depends on DnaC, implying that replication restart of blocked forks requires DnaB loading and possibly the entire assembly of new replisomes. Restart appears to occur synchronously when most lesions have been removed. This raises the possibility that restart and lesion removal are coupled. Both restart and cell division suffer long delays if lesion removal is prevented, but restart can occur. Our data fit well with models invoking the stalling of replication forks and their extensive processing before replication can restart. Delayed restart avoids the dangers of excessive recombination that might result if forks skipped over lesion after lesion, leaving many gaps in their wake.
Figures
References
-
- Arthur H.M., Lloyd R.G., Lloyd R.G. Hyper-recombination in uvrD mutants of Escherichia coli K-12. Mol. Gen. Genet. 1980;180:185–191. - PubMed
-
- Bierne H., Seigneur M., Ehrlich S.D., Michel B., Seigneur M., Ehrlich S.D., Michel B., Ehrlich S.D., Michel B., Michel B. uvrD mutations enhance tandem repeat deletion in the Escherichia coli chromosome via SOS induction of the RecF recombination pathway. Mol. Microbiol. 1997;26:557–567. - PubMed
-
- Camerini-Otero R.D., Hsieh P., Hsieh P. Parallel DNA triplexes, homologous recombination, and other homology-dependent DNA interactions. Cell. 1993;73:217–223. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
