Increases in the number of SNARE genes parallels the rise of multicellularity among the green plants
- PMID: 17369437
- PMCID: PMC1913785
- DOI: 10.1104/pp.106.092973
Increases in the number of SNARE genes parallels the rise of multicellularity among the green plants
Abstract
The green plant lineage is the second major multicellular expansion among the eukaryotes, arising from unicellular ancestors to produce the incredible diversity of morphologies and habitats observed today. In the unicellular ancestors, secretion of material through the endomembrane system was the major mechanism for interacting and shaping the external environment. In a multicellular organism, the external environment can be made of other cells, some of which may have vastly different developmental fates, or be part of different tissues or organs. In this context, a given cell must find ways to organize its secretory pathway at a level beyond that of the unicellular ancestor. Recently, sequence information from many green plants have become available, allowing an examination of the genomes for the machinery involved in the secretory pathway. In this work, the SNARE proteins of several green plants have been identified. While little increase in gene number was seen in the SNAREs of the early secretory system, many new SNARE genes and gene families have appeared in the multicellular green plants with respect to the unicellular plants, suggesting that this increase in the number of SNARE genes may have some relation to the rise of multicellularity in green plants.
Figures


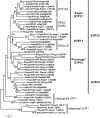
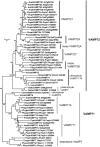

Similar articles
-
SNAREing the basis of multicellularity: consequences of protein family expansion during evolution.Mol Biol Evol. 2008 Sep;25(9):2055-68. doi: 10.1093/molbev/msn151. Epub 2008 Jul 10. Mol Biol Evol. 2008. PMID: 18621745
-
Differences in the SNARE evolution of fungi and metazoa.Biochem Soc Trans. 2009 Aug;37(Pt 4):787-91. doi: 10.1042/BST0370787. Biochem Soc Trans. 2009. PMID: 19614595 Review.
-
SNARE-ware: the role of SNARE-domain proteins in plant biology.Annu Rev Cell Dev Biol. 2007;23:147-74. doi: 10.1146/annurev.cellbio.23.090506.123529. Annu Rev Cell Dev Biol. 2007. PMID: 17506694 Review.
-
Comparative analysis of plant genomes allows the definition of the "Phytolongins": a novel non-SNARE longin domain protein family.BMC Genomics. 2009 Nov 4;10:510. doi: 10.1186/1471-2164-10-510. BMC Genomics. 2009. PMID: 19889231 Free PMC article.
-
Evolution of SET-domain protein families in the unicellular and multicellular Ascomycota fungi.BMC Evol Biol. 2008 Jul 1;8:190. doi: 10.1186/1471-2148-8-190. BMC Evol Biol. 2008. PMID: 18593478 Free PMC article.
Cited by
-
SNARE complexes of different composition jointly mediate membrane fusion in Arabidopsis cytokinesis.Mol Biol Cell. 2013 May;24(10):1593-601. doi: 10.1091/mbc.E13-02-0074. Epub 2013 Mar 20. Mol Biol Cell. 2013. PMID: 23515225 Free PMC article.
-
Conserved and plant-unique mechanisms regulating plant post-Golgi traffic.Front Plant Sci. 2012 Aug 28;3:197. doi: 10.3389/fpls.2012.00197. eCollection 2012. Front Plant Sci. 2012. PMID: 22973281 Free PMC article.
-
Qa-SNAREs localized to the trans-Golgi network regulate multiple transport pathways and extracellular disease resistance in plants.Proc Natl Acad Sci U S A. 2012 Jan 31;109(5):1784-9. doi: 10.1073/pnas.1115146109. Epub 2012 Jan 17. Proc Natl Acad Sci U S A. 2012. PMID: 22307646 Free PMC article.
-
SNAREs Regulate Vesicle Trafficking During Root Growth and Development.Front Plant Sci. 2022 Mar 14;13:853251. doi: 10.3389/fpls.2022.853251. eCollection 2022. Front Plant Sci. 2022. PMID: 35360325 Free PMC article. Review.
-
TaSYP137 and TaVAMP723, the SNAREs Proteins from Wheat, Reduce Resistance to Blumeria graminis f. sp. tritici.Int J Mol Sci. 2023 Mar 2;24(5):4830. doi: 10.3390/ijms24054830. Int J Mol Sci. 2023. PMID: 36902258 Free PMC article.
References
-
- Adl SM, Simpson AG, Farmer MA, Andersen RA, Anderson OR, Barta JR, Bowser SS, Brugerolle G, Fensome RA, Fredericq S, et al (2005) The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J Eukaryot Microbiol 52 399–451 - PubMed
-
- Bock JB, Matern HT, Peden AA, Scheller RH (2001) A genomic perspective on membrane compartment organization. Nature 409 839–841 - PubMed
-
- Burri L, Lithgow T (2004) A complete set of SNAREs in yeast. Traffic 5 45–52 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

