Oestrogen signalling inhibits invasive phenotype by repressing RelB and its target BCL2
- PMID: 17369819
- PMCID: PMC2394707
- DOI: 10.1038/ncb1559
Oestrogen signalling inhibits invasive phenotype by repressing RelB and its target BCL2
Abstract
Aberrant constitutive expression of c-Rel, p65 and p50 NF-kappaB subunits has been reported in over 90% of breast cancers. Recently, we characterized a de novo RelB NF-kappaB subunit synthesis pathway, induced by the cytomegalovirus (CMV) IE1 protein, in which binding of p50-p65 NF-kappaB and c-Jun-Fra-2 AP-1 complexes to the RELB promoter work in synergy to potently activate transcription. Although RelB complexes were observed in mouse mammary tumours induced by either ectopic c-Rel expression or carcinogen exposure, little is known about RelB in human breast disease. Here, we demonstrate constitutive de novo RelB synthesis is selectively active in invasive oestrogen receptor alpha (ERalpha)-negative breast cancer cells. ERalpha signalling reduced levels of functional NF-kappaB and Fra-2 AP-1 and inhibited de novo RelB synthesis, leading to an inverse correlation between RELB and ERalpha gene expression in human breast cancer tissues and cell lines. Induction of Bcl-2 by RelB promoted the more invasive phenotype of ERalpha-negative cancer cells. Thus, inhibition of de novo RelB synthesis represents a new mechanism whereby ERalpha controls epithelial to mesenchymal transition (EMT).
Figures



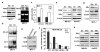
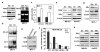



Comment in
-
NF-kappaB links oestrogen receptor signalling and EMT.Nat Cell Biol. 2007 Apr;9(4):361-3. doi: 10.1038/ncb0407-361. Nat Cell Biol. 2007. PMID: 17401385 No abstract available.
Similar articles
-
CD40 ligand-mediated activation of the de novo RelB NF-kappaB synthesis pathway in transformed B cells promotes rescue from apoptosis.J Biol Chem. 2007 Jun 15;282(24):17475-85. doi: 10.1074/jbc.M607313200. Epub 2007 Apr 19. J Biol Chem. 2007. PMID: 17446175
-
Induction of the RelB NF-kappaB subunit by the cytomegalovirus IE1 protein is mediated via Jun kinase and c-Jun/Fra-2 AP-1 complexes.J Virol. 2005 Jan;79(1):95-105. doi: 10.1128/JVI.79.1.95-105.2005. J Virol. 2005. PMID: 15596805 Free PMC article.
-
RelB NF-kappaB represses estrogen receptor alpha expression via induction of the zinc finger protein Blimp1.Mol Cell Biol. 2009 Jul;29(14):3832-44. doi: 10.1128/MCB.00032-09. Epub 2009 May 11. Mol Cell Biol. 2009. PMID: 19433448 Free PMC article.
-
RelB and Neuroinflammation.Cells. 2021 Jun 27;10(7):1609. doi: 10.3390/cells10071609. Cells. 2021. PMID: 34198987 Free PMC article. Review.
-
RelB: an outlier in leukocyte biology.J Leukoc Biol. 2013 Nov;94(5):941-51. doi: 10.1189/jlb.0513305. Epub 2013 Aug 6. J Leukoc Biol. 2013. PMID: 23922380 Free PMC article. Review.
Cited by
-
NFκB affects estrogen receptor expression and activity in breast cancer through multiple mechanisms.Mol Cell Endocrinol. 2015 Dec 15;418 Pt 3(0 3):235-9. doi: 10.1016/j.mce.2014.09.013. Epub 2014 Oct 18. Mol Cell Endocrinol. 2015. PMID: 25450861 Free PMC article. Review.
-
Panepoxydone targets NF-kB and FOXM1 to inhibit proliferation, induce apoptosis and reverse epithelial to mesenchymal transition in breast cancer.PLoS One. 2014 Jun 4;9(6):e98370. doi: 10.1371/journal.pone.0098370. eCollection 2014. PLoS One. 2014. Retraction in: PLoS One. 2023 Dec 29;18(12):e0296553. doi: 10.1371/journal.pone.0296553. PMID: 24896091 Free PMC article. Retracted.
-
Down-regulation of Forkhead box protein A1 (FOXA1) leads to cancer stem cell-like properties in tamoxifen-resistant breast cancer cells through induction of interleukin-6.J Biol Chem. 2017 May 19;292(20):8136-8148. doi: 10.1074/jbc.M116.763276. Epub 2017 Mar 7. J Biol Chem. 2017. PMID: 28270510 Free PMC article.
-
The NF-kappaB activation pathways, emerging molecular targets for cancer prevention and therapy.Expert Opin Ther Targets. 2010 Jan;14(1):45-55. doi: 10.1517/14728220903431069. Expert Opin Ther Targets. 2010. PMID: 20001209 Free PMC article. Review.
-
Multifaceted Interplay between Hormones, Growth Factors and Hypoxia in the Tumor Microenvironment.Cancers (Basel). 2022 Jan 21;14(3):539. doi: 10.3390/cancers14030539. Cancers (Basel). 2022. PMID: 35158804 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

