Review of the literature examining the correlation among DNA microarray technologies
- PMID: 17370338
- PMCID: PMC2682332
- DOI: 10.1002/em.20290
Review of the literature examining the correlation among DNA microarray technologies
Abstract
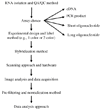
DNA microarray technologies are used in a variety of biological disciplines. The diversity of platforms and analytical methods employed has raised concerns over the reliability, reproducibility and correlation of data produced across the different approaches. Initial investigations (years 2000-2003) found discrepancies in the gene expression measures produced by different microarray technologies. Increasing knowledge and control of the factors that result in poor correlation among the technologies has led to much higher levels of correlation among more recent publications (years 2004 to present). Here, we review the studies examining the correlation among microarray technologies. We find that with improvements in the technology (optimization and standardization of methods, including data analysis) and annotation, analysis across platforms yields highly correlated and reproducible results. We suggest several key factors that should be controlled in comparing across technologies, and are good microarray practice in general.
Figures

References
-
- Ahmed FE. Microarray RNA transcriptional profiling. I. Platforms, experimental design and standardization. Expert Rev Mol Diagn. 2006a;6:535–550. - PubMed
-
- Ahmed FE. Microarray RNA transcriptional profiling. II. Analytical considerations and annotation. Expert Rev Mol Diagn. 2006b;6:703–715. - PubMed
-
- Andersen MT, Foy CA. The development of microarray standards. Anal Bioanal Chem. 2005;381:87–89. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

