Protein structure prediction by all-atom free-energy refinement
- PMID: 17371594
- PMCID: PMC1832197
- DOI: 10.1186/1472-6807-7-12
Protein structure prediction by all-atom free-energy refinement
Abstract
Background: The reliable prediction of protein tertiary structure from the amino acid sequence remains challenging even for small proteins. We have developed an all-atom free-energy protein forcefield (PFF01) that we could use to fold several small proteins from completely extended conformations. Because the computational cost of de-novo folding studies rises steeply with system size, this approach is unsuitable for structure prediction purposes. We therefore investigate here a low-cost free-energy relaxation protocol for protein structure prediction that combines heuristic methods for model generation with all-atom free-energy relaxation in PFF01.
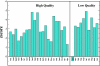
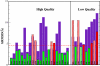
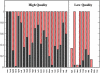
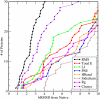
Results: We use PFF01 to rank and cluster the conformations for 32 proteins generated by ROSETTA. For 22/10 high-quality/low quality decoy sets we select near-native conformations with an average Calpha root mean square deviation of 3.03 A/6.04 A. The protocol incorporates an inherent reliability indicator that succeeds for 78% of the decoy sets. In over 90% of these cases near-native conformations are selected from the decoy set. This success rate is rationalized by the quality of the decoys and the selectivity of the PFF01 forcefield, which ranks near-native conformations an average 3.06 standard deviations below that of the relaxed decoys (Z-score).
Conclusion: All-atom free-energy relaxation with PFF01 emerges as a powerful low-cost approach toward generic de-novo protein structure prediction. The approach can be applied to large all-atom decoy sets of any origin and requires no preexisting structural information to identify the native conformation. The study provides evidence that a large class of proteins may be foldable by PFF01.
Figures



Similar articles
-
Template-free protein structure prediction and quality assessment with an all-atom free-energy model.Proteins. 2009 Nov 1;77(2):330-41. doi: 10.1002/prot.22438. Proteins. 2009. PMID: 19422063
-
Improved protein structure selection using decoy-dependent discriminatory functions.BMC Struct Biol. 2004 Jun 18;4:8. doi: 10.1186/1472-6807-4-8. BMC Struct Biol. 2004. PMID: 15207004 Free PMC article.
-
Efficient identification of near-native conformations in ab initio protein structure prediction using structural profiles.Proteins. 2010 Feb 1;78(2):249-58. doi: 10.1002/prot.22533. Proteins. 2010. PMID: 19701942
-
Protein decoy sets for evaluating energy functions.J Biomol Struct Dyn. 2004 Jun;21(6):725-36. doi: 10.1080/07391102.2004.10506963. J Biomol Struct Dyn. 2004. PMID: 15106995 Review.
-
Methods for the Refinement of Protein Structure 3D Models.Int J Mol Sci. 2019 May 9;20(9):2301. doi: 10.3390/ijms20092301. Int J Mol Sci. 2019. PMID: 31075942 Free PMC article. Review.
Cited by
-
Computational protein structure refinement: Almost there, yet still so far to go.Wiley Interdiscip Rev Comput Mol Sci. 2017 May-Jun;7(3):e1307. doi: 10.1002/wcms.1307. Epub 2017 Mar 28. Wiley Interdiscip Rev Comput Mol Sci. 2017. PMID: 30613211 Free PMC article.
-
Unsupervised and Supervised Learning over theEnergy Landscape for Protein Decoy Selection.Biomolecules. 2019 Oct 14;9(10):607. doi: 10.3390/biom9100607. Biomolecules. 2019. PMID: 31615116 Free PMC article.
-
Use of decoys to optimize an all-atom force field including hydration.Biophys J. 2008 Sep;95(5):2434-49. doi: 10.1529/biophysj.108.133587. Epub 2008 May 23. Biophys J. 2008. PMID: 18502794 Free PMC article.
-
Identifying native-like protein structures with scoring functions based on all-atom ECEPP force fields, implicit solvent models and structure relaxation.Proteins. 2009 Oct;77(1):38-51. doi: 10.1002/prot.22414. Proteins. 2009. PMID: 19384995 Free PMC article.
-
Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples.Proc Natl Acad Sci U S A. 2012 Jul 3;109(27):10873-8. doi: 10.1073/pnas.1203013109. Epub 2012 Jun 25. Proc Natl Acad Sci U S A. 2012. PMID: 22733734 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

